1YU3
 
 | | Major Tropism Determinant I1 Variant | | Descriptor: | MAGNESIUM ION, Major Tropism Determinant (Mtd-I1) | | Authors: | McMahon, S.A, Miller, J.L, Lawton, J.A, Ghosh, P. | | Deposit date: | 2005-02-11 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The C-type lectin fold as an evolutionary solution for massive sequence variation
Nat.Struct.Mol.Biol., 12, 2005
|
|
4MP4
 
 | | Crystal structure of a glutathione transferase family member from Acinetobacter baumannii, Target EFI-501785, apo structure | | Descriptor: | Glutathione S-transferase, SULFATE ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-12 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Acinetobacter baumannii, Target EFI-501785, apo structure
To be Published
|
|
4D2B
 
 | | Structure of a ligand free POT family peptide transporter | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, DI-OR TRIPEPTIDE:H+ SYMPORTER, ... | | Authors: | Lyons, J.A, Parker, J.L, Solcan, N, Brinth, A, Li, D, Shah, S.T.A, Caffrey, M, Newstead, S. | | Deposit date: | 2014-05-09 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for Polyspecificity in the Pot Family of Proton-Coupled Oligopeptide Transporters.
Embo Rep., 15, 2014
|
|
4MNC
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_4736), Target EFI-510156, with bound benzoyl formate, space group P21 | | Descriptor: | BENZOYL-FORMIC ACID, SULFATE ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-10 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
1FBQ
 
 | |
1FQC
 
 | | CRYSTAL STRUCTURE OF MALTOTRIOTOL BOUND TO CLOSED-FORM MALTODEXTRIN BINDING PROTEIN | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-D-glucose | | Authors: | Duan, X, Hall, J.A, Nikaido, H, Quiocho, F.A. | | Deposit date: | 2000-09-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding.
J.Mol.Biol., 306, 2001
|
|
1FQL
 
 | | X-RAY CRYSTAL STRUCTURE OF ZINC-BOUND F95M/W97V CARBONIC ANHYDRASE (CAII) VARIANT | | Descriptor: | CARBONIC ANHYDRASE, MERCURY (II) ION, ZINC ION | | Authors: | Cox, J.D, Hunt, J.A, Compher, K.M, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2000-09-05 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural influence of hydrophobic core residues on metal binding and specificity in carbonic anhydrase II.
Biochemistry, 39, 2000
|
|
1ZUQ
 
 | | Contribution to Structure and Catalysis of Tyrosine 34 in Human Manganese Superoxide Dismutase | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Hearn, A.S, Perry, J.J, Cabelii, D.E, Tainer, J.A, Nick, H.S, Silverman, D.S. | | Deposit date: | 2005-05-31 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of human manganese superoxide dismutase tyrosine 34 to structure and catalysis.
Biochemistry, 48, 2009
|
|
1FL7
 
 | | HUMAN FOLLICLE STIMULATING HORMONE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLLICLE STIMULATING PROTEIN ALPHA CHAIN, ... | | Authors: | Fox, K.M, Dias, J.A, Van Roey, P. | | Deposit date: | 2000-08-11 | | Release date: | 2001-03-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structure of human follicle-stimulating hormone.
Mol.Endocrinol., 15, 2001
|
|
4MPI
 
 | | Crystal structure of the chitin-binding module (CBM18) of a chitinase-like protein from Hevea brasiliensis | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Class I chitinase | | Authors: | Martinez-Caballero, C.S, Hermoso, J.A, Rodriguez-Romero, A. | | Deposit date: | 2013-09-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Comparative study of two GH19 chitinase-like proteins from Hevea brasiliensis, one exhibiting a novel carbohydrate-binding domain.
Febs J., 281, 2014
|
|
1ZSP
 
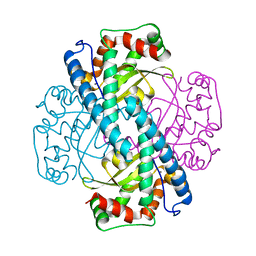 | | Contribution to Structure and Catalysis of Tyrosine 34 in Human Manganese Superoxide Dismutase | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Hearn, A.S, Perry, J.J, Cabelli, D.E, Tainer, J.A, Nick, H.S, Silverman, D.S. | | Deposit date: | 2005-05-24 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Contribution of human manganese superoxide dismutase tyrosine 34 to structure and catalysis.
Biochemistry, 48, 2009
|
|
4C97
 
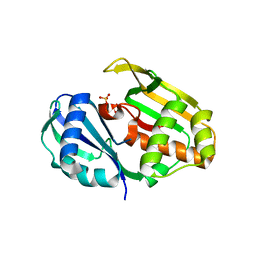 | | Cas6 (TTHA0078) H37A mutant | | Descriptor: | CAS6A, SULFATE ION | | Authors: | Jinek, M, Niewoehner, O, Doudna, J.A. | | Deposit date: | 2013-10-02 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of Crispr RNA Recognition and Processing by Cas6 Endonucleases.
Nucleic Acids Res., 42, 2014
|
|
4C61
 
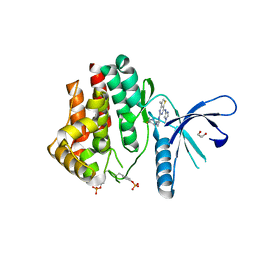 | | Inhibitors of Jak2 Kinase domain | | Descriptor: | ACETATE ION, N2-[(1S)-1-(5-fluoropyrimidin-2-yl)ethyl]-7-methyl-N4-(1-methylimidazol-4-yl)thieno[3,2-d]pyrimidine-2,4-diamine, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Read, J.A, Green, I, Pollard, H, Howard, T. | | Deposit date: | 2013-09-17 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of 1-Methyl-1H-Imidazole Derivatives as Potent Jak2 Inhibitors.
J.Med.Chem., 57, 2014
|
|
4C6H
 
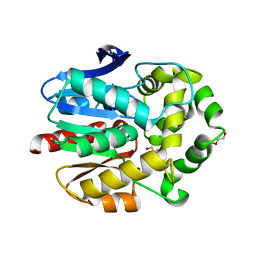 | | Haloalkane dehalogenase with 1-hexanol | | Descriptor: | CHLORIDE ION, HALOALKANE DEHALOGENASE, HEXAN-1-OL, ... | | Authors: | Novak, H.R, Sayer, C, Isupov, M, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Biochemical and Structural Characterisation of a Haloalkane Dehalogenase from a Marine Rhodobacteraceae.
FEBS Lett., 588, 2014
|
|
1ZRB
 
 | | Thrombin in complex with an azafluorenyl inhibitor 23b | | Descriptor: | 11-peptide hirudin fragment, 3-AZA-9-HYDROXY-9-FLUORENYLCARBONYL-L-PROLYL-2-AMINOMETHYL-5-CHLOROBENZYLAMIDE, N-OXIDE, ... | | Authors: | Stauffer, K.J, Williams, P.D, Selnick, H.G, Nantermet, P.G, Newton, C.L, Homnick, C.F, Zrada, M.M, Lewis, S.D, Lucas, B.J, Krueger, J.A, Pietrak, B.L, Lyle, E.A, Singh, R, Miller-Stein, C, White, R.B, Wong, B, Wallace, A.A, Sitko, G.R, Cook, J.J, Holahan, M.A, Stranieri-Michener, M, Leonard, Y.M, Lynch Jr, J.J, McMasters, D.R, Yan, Y. | | Deposit date: | 2005-05-19 | | Release date: | 2005-06-07 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 9-hydroxyazafluorenes and their use in thrombin inhibitors
J.Med.Chem., 48, 2005
|
|
4ETL
 
 | | Crystallographic structure of phenylalanine hydroxylase from Chromobacterium violaceum F258A mutation | | Descriptor: | COBALT (II) ION, Phenylalanine-4-hydroxylase | | Authors: | Ronau, J.A, Paul, L.P, Corn, I.R, Wagner, K.T, Abu-Omar, M.M, Das, C. | | Deposit date: | 2012-04-24 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | An additional substrate binding site in a bacterial phenylalanine hydroxylase.
Eur.Biophys.J., 42, 2013
|
|
4EXJ
 
 | | Crystal structure of glutathione s-transferase like protein lelg_03239 (target efi-501752) from lodderomyces elongisporus | | Descriptor: | CHLORIDE ION, SULFATE ION, uncharacterized protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of glutathione s-transferase like protein lelg_03239 (target efi-501752) from lodderomyces elongisporus
To be Published
|
|
4MST
 
 | |
1GWF
 
 | | Compound II structure of Micrococcus Lysodeikticus catalase | | Descriptor: | ACETATE ION, Catalase, OXYGEN ATOM, ... | | Authors: | Murshudov, G.N, Grebenko, A.I, Brannigan, J.A, Antson, A.A, Barynin, V.V, Dodson, G.G, Dauter, Z, Wilson, K.S, Melik-Adamyan, W.R. | | Deposit date: | 2002-03-15 | | Release date: | 2002-04-26 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Structures of Micrococcus Lysodeikticus Catalase, its Ferryl Intermediate (Compound II) and Nadph Complex.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GNO
 
 | | HIV-1 PROTEASE (WILD TYPE) COMPLEXED WITH U89360E (INHIBITOR) | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Treharne, A, Hartsuck, J.A, Foundling, S, Tang, J. | | Deposit date: | 1996-05-04 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of complexes of a peptidic inhibitor with wild-type and two mutant HIV-1 proteases.
Biochemistry, 35, 1996
|
|
2A8I
 
 | | Crystal Structure of human Taspase1 | | Descriptor: | Threonine aspartase 1 | | Authors: | Khan, J.A, Dunn, B.M, Tong, L. | | Deposit date: | 2005-07-08 | | Release date: | 2005-11-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Taspase1, a Crucial Protease Regulating the Function of MLL.
Structure, 13, 2005
|
|
2AAX
 
 | | Mineralocorticoid Receptor Double Mutant with Bound Cortisone | | Descriptor: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, Mineralocorticoid receptor, SULFATE ION | | Authors: | Bledsoe, R.K, Madauss, K.P, Holt, J.A, Apolito, C.J, Lambert, M.H, Pearce, K.H, Stanley, T.B, Stewart, E.L, Trump, R.P, Willson, T.M, Williams, S.P. | | Deposit date: | 2005-07-14 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Ligand-mediated Hydrogen Bond Network Required for the Activation of the Mineralocorticoid Receptor
J.Biol.Chem., 280, 2005
|
|
4N15
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Burkholderia ambifaria (BAM_6123), Target EFI-510059, with bound beta-D-glucuronate | | Descriptor: | MAGNESIUM ION, TRAP dicarboxylate transporter, DctP subunit, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Zhao, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N2Y
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Archaeoglobus fulgidus | | Descriptor: | GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-10-06 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Archaeoglobus fulgidus
To be Published
|
|
1GUM
 
 | | HUMAN GLUTATHIONE TRANSFERASE A4-4 WITHOUT LIGANDS | | Descriptor: | PROTEIN (GLUTATHIONE TRANSFERASE A4-4) | | Authors: | Bruns, C.M, Hubatsch, I, Ridderstrom, M, Mannervik, B, Tainer, J.A. | | Deposit date: | 1998-06-11 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human glutathione transferase A4-4 crystal structures and mutagenesis reveal the basis of high catalytic efficiency with toxic lipid peroxidation products
J.Mol.Biol., 288, 1999
|
|
