8R8T
 
 | |
1QJX
 
 | | HUMAN RHINOVIRUS 16 COAT PROTEIN IN COMPLEX WITH ANTIVIRAL COMPOUND WIN68934 | | Descriptor: | 2,6-DIMETHYL-1-(3-[3-METHYL-5-ISOXAZOLYL]-PROPANYL)-4-[4-METHYL-2H-TETRAZOL-2-YL]-PHENOL, MYRISTIC ACID, PROTEIN VP1, ... | | Authors: | Hadfield, A.T, Diana, G.D, Rossmann, M.G. | | Deposit date: | 1999-07-06 | | Release date: | 1999-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of Three Structurally Related Antiviral Compounds in Complex with Human Rhinovirus 16
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2OE9
 
 | | High-pressure structure of pseudo-WT T4 Lysozyme | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2006-12-28 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Rigidity of a Large Cavity-containing Protein Revealed by High-pressure Crystallography.
J.Mol.Biol., 367, 2007
|
|
2OE7
 
 | | High-Pressure T4 Lysozyme | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2006-12-28 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Rigidity of a Large Cavity-containing Protein Revealed by High-pressure Crystallography.
J.Mol.Biol., 367, 2007
|
|
1QJU
 
 | | HUMAN RHINOVIRUS 16 COAT PROTEIN IN COMPLEX WITH ANTIVIRAL COMPOUND VP61209 | | Descriptor: | 2,6-DIMETHYL-1-(3-[3-METHYL-5-ISOXAZOLYL]-PROPANYL)-4-[2N-METHYL-2H-TETRAZOL-5-YL]-PHENOL, MYRISTIC ACID, PROTEIN VP1, ... | | Authors: | Hadfield, A.T, Minor, I, Diana, G.D, Rossmann, M.G. | | Deposit date: | 1999-07-05 | | Release date: | 1999-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of Three Structurally Related Antiviral Compounds in Complex with Human Rhinovirus 16
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QJY
 
 | | HUMAN RHINOVIRUS 16 COAT PROTEIN IN COMPLEX WITH ANTIVIRAL COMPOUND VP65099 | | Descriptor: | 2,6-DIMETHYL-1-(3-[3-METHYL-5-ISOXAZOLYL]-PROPANYL)-4-[2-METHYL-4-ISOXAZOLYL]-PHENOL, MYRISTIC ACID, PROTEIN VP1, ... | | Authors: | Hadfield, A.T, Diana, G.D, Rossmann, M.G. | | Deposit date: | 1999-07-06 | | Release date: | 1999-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of Three Structurally Related Antiviral Compounds in Complex with Human Rhinovirus 16
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2OEA
 
 | | High-pressure structure of pseudo-WT T4 Lysozyme | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2006-12-28 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Rigidity of a Large Cavity-containing Protein Revealed by High-pressure Crystallography.
J.Mol.Biol., 367, 2007
|
|
4MLB
 
 | | Reverse polarity of binding pocket suggests different function of a MOP superfamily transporter from Pyrococcus furiosus Vc1 (DSM3638) | | Descriptor: | CHLORIDE ION, PENTAETHYLENE GLYCOL MONODECYL ETHER, PF0708 | | Authors: | Malviya, V.N, Nonaka, T, Muenke, C, Koepke, J, Michel, H. | | Deposit date: | 2013-09-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Inward-facing conformation of a multidrug resistance MATE family transporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1MEC
 
 | |
2ITG
 
 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE: ORDERED ACTIVE SITE IN THE F185H CONSTRUCT | | Descriptor: | HUMAN IMMUNODEFICIENCY VIRUS-1 INTEGRASE | | Authors: | Bujacz, G, Alexandratos, J, Wlodawer, A, Zhou-Liu, Q, Clement-Mella, C. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The catalytic domain of human immunodeficiency virus integrase: ordered active site in the F185H mutant.
FEBS Lett., 398, 1996
|
|
6VK3
 
 | | CryoEM structure of Hrd3/Yos9 complex | | Descriptor: | Hrd3, Protein OS-9 homolog | | Authors: | Wu, X, Rapoport, T.A. | | Deposit date: | 2020-01-18 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
6VJZ
 
 | | CryoEM structure of Hrd1-Usa1/Der1/Hrd3 complex of the expected topology | | Descriptor: | Degradation in the endoplasmic reticulum protein 1, ERAD-associated E3 ubiquitin-protein ligase HRD1, ERAD-associated E3 ubiquitin-protein ligase component HRD3, ... | | Authors: | Wu, X, Rapoport, T.A. | | Deposit date: | 2020-01-18 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
6VK1
 
 | | CryoEM structure of Hrd1/Hrd3 part from Hrd1-Usa1/Der1/Hrd3 complex | | Descriptor: | ERAD-associated E3 ubiquitin-protein ligase HRD1, ERAD-associated E3 ubiquitin-protein ligase component HRD3 | | Authors: | Wu, X, Rapoport, T.A. | | Deposit date: | 2020-01-18 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
1RHI
 
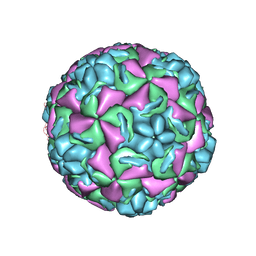 | | HUMAN RHINOVIRUS 3 COAT PROTEIN | | Descriptor: | CALCIUM ION, HUMAN RHINOVIRUS 3 COAT PROTEIN | | Authors: | Zhao, R, Rossmann, M.G. | | Deposit date: | 1996-06-17 | | Release date: | 1997-03-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human rhinovirus 3 at 3.0 A resolution.
Structure, 4, 1996
|
|
6VK0
 
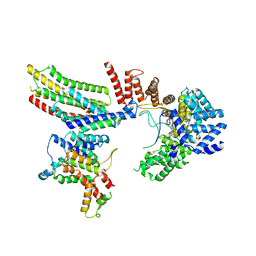 | | CryoEM structure of Hrd1-Usa1/Der1/Hrd3 of the flipped topology | | Descriptor: | Degradation in the endoplasmic reticulum protein 1, ERAD-associated E3 ubiquitin-protein ligase HRD1, ERAD-associated E3 ubiquitin-protein ligase component HRD3, ... | | Authors: | Wu, X, Rapoport, T.A. | | Deposit date: | 2020-01-18 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
3NDB
 
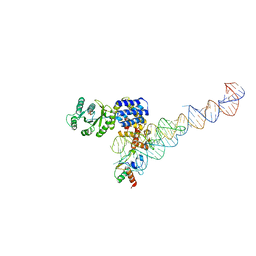 | | Crystal structure of a signal sequence bound to the signal recognition particle | | Descriptor: | PHOSPHATE ION, SRP RNA, Signal recognition 54 kDa protein, ... | | Authors: | Hainzl, T, Huang, S, Sauer-Eriksson, E. | | Deposit date: | 2010-06-07 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of signal-sequence recognition by the signal recognition particle.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3PFW
 
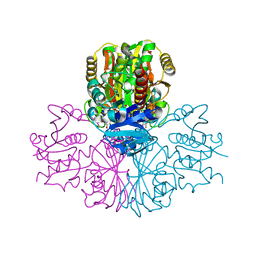 | | Crystal structure of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase (GAPDS) complex with NAD, a binary form | | Descriptor: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, testis-specific, ... | | Authors: | Chaikuad, A, Shafqat, N, Yue, W.W, Cocking, R, Bray, J.E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-29 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and kinetic characterization of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase, GAPDS.
Biochem.J., 435, 2011
|
|
6VJY
 
 | | Cryo-EM structure of Hrd1/Hrd3 monomer | | Descriptor: | ERAD-associated E3 ubiquitin-protein ligase HRD1, ERAD-associated E3 ubiquitin-protein ligase component HRD3 | | Authors: | Wu, X, Rapoport, T.A. | | Deposit date: | 2020-01-18 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
5OKL
 
 | | Human afamin monoclinic crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Afamin, ... | | Authors: | Rupp, B, Naschberger, A, Bowler, M.W. | | Deposit date: | 2017-07-25 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Evidence for a Role of the Multi-functional Human Glycoprotein Afamin in Wnt Transport.
Structure, 25, 2017
|
|
2PJW
 
 | |
7NL7
 
 | | Crystal Structure of DC-SIGN in complex with a triazole-based glycomimetic ligand | | Descriptor: | 3-Aminopropyl 2-deoxy-2-(4-phenyl-1,2,3-triazol-1-yl)-alpha-D-mannopyranoside, CALCIUM ION, DC-SIGN, ... | | Authors: | Jakob, R.P, Cramer, J, Lakkaichi, A, Aliu, B, Cattaneo, I, Klein, S, Jiang, X, Rabbani, S, Schwardt, O, Ernst, B, Maier, T. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sweet Drugs for Bad Bugs: A Glycomimetic Strategy against the DC-SIGN-Mediated Dissemination of SARS-CoV-2.
J.Am.Chem.Soc., 143, 2021
|
|
7NL6
 
 | | Crystal Structure of DC-SIGN in complex with a triazole-based glycomimetic ligand | | Descriptor: | CALCIUM ION, DC-SIGN, CRD domain, ... | | Authors: | Jakob, R.P, Cramer, J, Lakkaichi, A, Aliu, B, Cattaneo, I, Klein, S, Jiang, X, Rabbani, S, Schwardt, O, Ernst, B, Maier, T. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sweet Drugs for Bad Bugs: A Glycomimetic Strategy against the DC-SIGN-Mediated Dissemination of SARS-CoV-2.
J.Am.Chem.Soc., 143, 2021
|
|
1RUS
 
 | | CRYSTAL STRUCTURE OF THE BINARY COMPLEX OF RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE AND ITS PRODUCT, 3-PHOSPHO-D-GLYCERATE | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, RUBISCO (RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE) | | Authors: | Lundqvist, T, Schneider, G. | | Deposit date: | 1991-10-10 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the binary complex of ribulose-1,5-bisphosphate carboxylase and its product, 3-phospho-D-glycerate.
J.Biol.Chem., 264, 1989
|
|
1VRH
 
 | | HRV14/SDZ 880-061 COMPLEX | | Descriptor: | 2-[4-(2H-1,4-BENZOTHIAZINE-3-YL)-PIPERAZINE-1-LY]-1,3-THIAZOLE-4-CARBOXYLIC ACID ETHYLESTER, RHINOVIRUS 14 | | Authors: | Oren, D.A, Zhang, A, Arnold, E. | | Deposit date: | 1996-02-26 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis and activity of piperazine-containing antirhinoviral agents and crystal structure of SDZ 880-061 bound to human rhinovirus 14.
J.Mol.Biol., 259, 1996
|
|
8QYQ
 
 | | Beta-cardiac myosin S1 fragment in the pre-powerstroke state complexed to Mavacamten | | Descriptor: | 6-[[(1~{S})-1-phenylethyl]amino]-3-propan-2-yl-1~{H}-pyrimidine-2,4-dione, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Robert-Paganin, J, Kikuti, C, Auguin, D, Rety, S, David, A, Houdusse, A. | | Deposit date: | 2023-10-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Omecamtiv mecarbil and Mavacamten target the same myosin pocket despite antagonistic effects in heart contraction.
Biorxiv, 2023
|
|
