6CHO
 
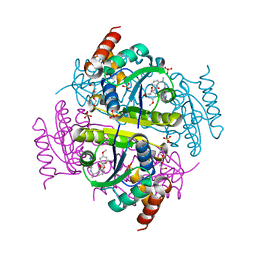 | | Phosphopantetheine adenylyltransferase (CoaD) in complex with (R)-2-((1-(3-(4-methoxyphenoxy)phenyl)ethyl)amino)-5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one | | Descriptor: | 2-({(1R)-1-[3-(4-methoxyphenoxy)phenyl]ethyl}amino)-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7(6H)-one, Phosphopantetheine adenylyltransferase, SULFATE ION, ... | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-22 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Optimization of Phosphopantetheine Adenylyltransferase Inhibitors with Gram-Negative Antibacterial Activity.
J. Med. Chem., 61, 2018
|
|
7UYV
 
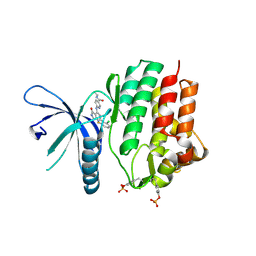 | | Crystal structure of JAK3 kinase domain in complex with compound 25 | | Descriptor: | 6-{[(2M)-2-(2-chloro-6-fluorophenyl)-5-oxo-5H-pyrrolo[3,4-b]pyridin-4-yl]amino}-N-ethylpyridine-3-carboxamide, CHLORIDE ION, Tyrosine-protein kinase JAK3 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYS
 
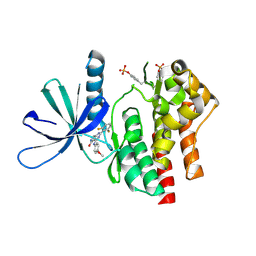 | | Crystal structure of TYK2 kinase domain in complex with compound 16 | | Descriptor: | 2-(2,6-difluorophenyl)-4-(4-methoxyanilino)-5H-pyrrolo[3,4-d]pyrimidin-5-one, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
6CKW
 
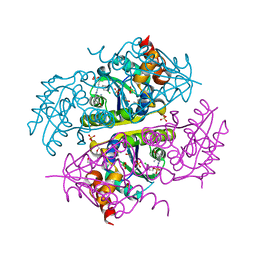 | | Phosphopantetheine adenylyltransferase (CoaD) in complex with (R)-3-((7-(((S)-2-amino-2-(2-methoxyphenyl)ethyl)amino)-5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)amino)-3-(3-chlorophenyl)propanenitrile | | Descriptor: | (3R)-3-[(7-{[(2S)-2-amino-2-(2-methoxyphenyl)ethyl]amino}-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)amino]-3-(3-chlorophenyl)propanenitrile, DIMETHYL SULFOXIDE, Phosphopantetheine adenylyltransferase, ... | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-03-01 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Discovery and Optimization of Phosphopantetheine Adenylyltransferase Inhibitors with Gram-Negative Antibacterial Activity.
J. Med. Chem., 61, 2018
|
|
7UHM
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Cleaved Moxalactam (150 ms Snapshot) | | Descriptor: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7JX3
 
 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab domain of monoclonal antibody S2H14, Heavy chain of Fab domain of monoclonal antibody S304, ... | | Authors: | Snell, G, Czudnochowski, N, Rosen, L.E, Nix, J.C, Corti, D, Veesler, D, Park, Y.J, Walls, A.C, Tortorici, M.A, Cameroni, E, Pinto, D, Beltramello, M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-08-26 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
6VKL
 
 | | Negative stain reconstruction of the yeast exocyst octameric complex. | | Descriptor: | Exocyst complex component EXO70, Exocyst complex component EXO84, Exocyst complex component SEC10, ... | | Authors: | Frost, A, Munson, M. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Exocyst structural changes associated with activation of tethering downstream of Rho/Cdc42 GTPases.
J. Cell Biol., 219, 2020
|
|
7P11
 
 | | Galectin-8 N-terminal carbohydrate recognition domain in complex with quinoline D-galactal ligand | | Descriptor: | 2-[[(2~{R},3~{R},4~{R})-2-(hydroxymethyl)-3-oxidanyl-3,4-dihydro-2~{H}-pyran-4-yl]oxymethyl]quinoline-7-carboxylic acid, CHLORIDE ION, Galectin-8, ... | | Authors: | Hassan, M, Hakansson, M, Nilsson, J.U, Kovacic, R. | | Deposit date: | 2021-07-01 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design of d-Galactal Derivatives with High Affinity and Selectivity for the Galectin-8 N-Terminal Domain.
Acs Med.Chem.Lett., 12, 2021
|
|
6D2L
 
 | | Crystal structure of human CARM1 with (S)-SKI-72 | | Descriptor: | (2S,5S)-2-amino-6-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-5-[(benzylamino)methyl]-N-[2-(4-hydroxyphenyl)ethyl]hexanamide, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | DONG, A, ZENG, H, WALKER, J.R, Hutchinson, A, Seitova, A, LUO, M, CAI, X.C, KE, W, WANG, J, SHI, C, ZHENG, W, LEE, J.P, IBANEZ, G, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-13 | | Release date: | 2018-05-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A chemical probe of CARM1 alters epigenetic plasticity against breast cancer cell invasion.
Elife, 8, 2019
|
|
6QEG
 
 | | CRYSTAL STRUCTURE OF HUMAN METHIONINE AMINOPEPTIDASE-2 IN COMPLEX WITH AN INHIBITOR 2-Oxo-1-phenyl-pyrrolidine-3-carboxylic acid (2-thiophen-2-yl-ethyl)-amide | | Descriptor: | (3~{S})-3-oxidanyl-2-oxidanylidene-1-phenyl-~{N}-(2-thiophen-2-ylethyl)pyrrolidine-3-carboxamide, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Musil, D, Heinrich, T, Lehmann, M. | | Deposit date: | 2019-01-07 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery and Structure-Based Optimization of Next-Generation Reversible Methionine Aminopeptidase-2 (MetAP-2) Inhibitors.
J.Med.Chem., 62, 2019
|
|
6CEC
 
 | | Crystal structure of fragment 3-(3-Methoxy-2-quinoxalinyl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(3-methoxyquinoxalin-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Franzoni, I, Ravichandran, M, Lautens, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
7P1M
 
 | | Galectin-8 N-terminal carbohydrate recognition domain in complex with benzimidazole D-galactal ligand | | Descriptor: | 2-[[(2R,3R,4R)-2-(hydroxymethyl)-3-oxidanyl-3,4-dihydro-2H-pyran-4-yl]oxymethyl]-3-methyl-benzimidazole-5-carboxylic acid, CHLORIDE ION, Galectin-8 | | Authors: | Hassan, M, Hakansson, M, Nilsson, J.U, Kovacic, R. | | Deposit date: | 2021-07-02 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure-Guided Design of d-Galactal Derivatives with High Affinity and Selectivity for the Galectin-8 N-Terminal Domain.
Acs Med.Chem.Lett., 12, 2021
|
|
7UHO
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Cleaved Moxalactam (500 ms Snapshot) | | Descriptor: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
5N15
 
 | | First Bromodomain (BD1) from Candida albicans Bdf1 in the unbound form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bromodomain-containing factor 1, GLYCEROL, ... | | Authors: | Mietton, F, Ferri, E, Champleboux, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvel, M, d'Enfert, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | Deposit date: | 2017-02-05 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
7OPQ
 
 | | Rab27a fusion with Slp2a-RBDa1 effector covalent adduct with CA1 in C188 | | Descriptor: | 1-[(2~{S})-2-(4-methoxyphenyl)pyrrolidin-1-yl]propan-1-one, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Jamshidiha, M, Tersa, M, Lanyon-Hogg, T, Perez-Dorado, I, Sutherell, C.L, De Vita, E, Morgan, R.M.L, Tate, E.W, Cota, E. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Identification of the first structurally validated covalent ligands of the small GTPase RAB27A.
Rsc Med Chem, 13, 2022
|
|
6Z35
 
 | | De-novo Maquette 2 protein with buried ion-pair | | Descriptor: | Maquette 2-1ip | | Authors: | Baumgart, M, Roepke, M, Muehlbauer, M, Asami, S, Mader, S, Fredriksson, K, Groll, M, Gamiz-Hernandez, A.P, Kaila, V.R.I. | | Deposit date: | 2020-05-19 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Design of buried charged networks in artificial proteins.
Nat Commun, 12, 2021
|
|
7OPR
 
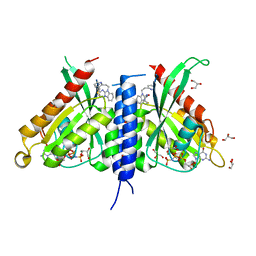 | | Rab27a fusion with Slp2a-RBDa1 effector covalent adduct with CB1 in C123 | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Jamshidiha, M, Tersa, M, Lanyon-Hogg, T, Perez-Dorado, I, Sutherell, C.L, De Vita, E, Morgan, R.M.L, Tate, E.W, Cota, E. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Identification of the first structurally validated covalent ligands of the small GTPase RAB27A.
Rsc Med Chem, 13, 2022
|
|
7OPP
 
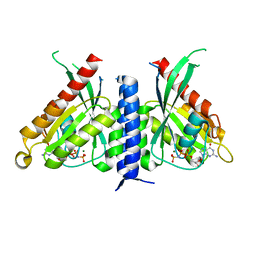 | | Crystal structure of the Rab27a fusion with Slp2a-RBDa1 effector for SF4 pocket drug targeting | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Synaptotagmin-like protein 2,Ras-related protein Rab-27A | | Authors: | Jamshidiha, M, Tersa, M, Lanyon-Hogg, T, Perez-Dorado, I, Sutherell, C.L, De Vita, E, Morgan, R.M.L, Tate, E.W, Cota, E. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Identification of the first structurally validated covalent ligands of the small GTPase RAB27A.
Rsc Med Chem, 13, 2022
|
|
5FVG
 
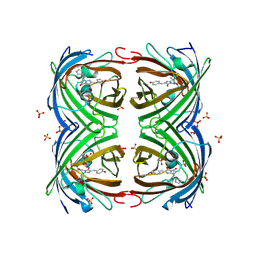 | | Structure of IrisFP at 100 K. | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Colletier, J.P, Gallat, F.X, Coquelle, N, Weik, M. | | Deposit date: | 2016-02-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein Irisfp.
J.Phys.Chem.Lett, 7, 2016
|
|
5WS6
 
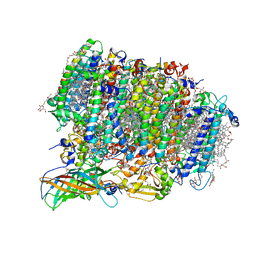 | | Native XFEL structure of Photosystem II (preflash two-flash dataset | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Light-induced structural changes and the site of O=O bond formation in PSII caught by XFEL.
Nature, 543, 2017
|
|
6CHP
 
 | |
6WX7
 
 | | SOX2 bound to Importin-alpha 2 | | Descriptor: | Importin subunit alpha-1, Transcription factor SOX-2 | | Authors: | Bikshapathi, J, Stewart, M, Forwood, J.K, Aragao, D, Roman, N. | | Deposit date: | 2020-05-09 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for nuclear import selectivity of pioneer transcription factor SOX2.
Nat Commun, 12, 2021
|
|
7OY9
 
 | |
5KDD
 
 | | Apo-structure of humanised RadA-mutant humRadA22 | | Descriptor: | DNA repair and recombination protein RadA, SULFATE ION | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
6BRB
 
 | | Novel non-antibody protein scaffold targeting CD40L | | Descriptor: | CD40 ligand, Tn3-like, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Oganesyan, V, Baca, M, Thisted, T, Grinberg, L, Wu, H, Dall'Acqua, W.F. | | Deposit date: | 2017-11-30 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | A CD40L-targeting protein reduces autoantibodies and improves disease activity in patients with autoimmunity.
Sci Transl Med, 11, 2019
|
|
