4FZZ
 
 | | Exonuclease X in complex with 5' overhanging duplex DNA | | Descriptor: | DNA (5'-D(*GP*TP*CP*AP*TP*TP*GP*TP*GP*GP*AP*TP*CP*CP*GP*AP*G)-3'), Exodeoxyribonuclease 10, SODIUM ION | | Authors: | Wang, T, Sun, H, Cheng, F, Bi, L, Jiang, T. | | Deposit date: | 2012-07-08 | | Release date: | 2013-07-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recognition and processing of double-stranded DNA by ExoX, a distributive 3'-5' exonuclease
Nucleic Acids Res., 41, 2013
|
|
7XRP
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with nanobody C5G2 (localized refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C5G2 nanobody, Spike protein S1 | | Authors: | Liu, L, Sun, H, Jiang, Y, Liu, X, Zhao, D, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-05-11 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | A potent synthetic nanobody with broad-spectrum activity neutralizes SARS-CoV-2 virus and the Omicron variant BA.1 through a unique binding mode.
J Nanobiotechnology, 20, 2022
|
|
8QB3
 
 | | ADDobody zinc containing condition | | Descriptor: | ADDobody, ZINC ION | | Authors: | Buzas, D, Toelzer, C, Berger, I, Schaffitzel, C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
8QBX
 
 | | Chimeric Adenovirus-derived dodecamer | | Descriptor: | Penton protein | | Authors: | Buzas, D, Borucu, U, Bufton, J, Kapadalakere, S.Y, Toelzer, C. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
5TKB
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 4D IN COMPLEX WITH A TETRAFLUORANLINE COMPOUND | | Descriptor: | ETHANOL, MAGNESIUM ION, N-[(2R)-2,3-dihydroxy-2-methylpropyl]-8-(methylamino)-6-[(2,3,5,6-tetrafluorophenyl)amino]imidazo[1,2-b]pyridazine-3-carboxamide, ... | | Authors: | Sack, J.S. | | Deposit date: | 2016-10-06 | | Release date: | 2016-12-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Identification of imidazo[1,2-b]pyridazine TYK2 pseudokinase ligands as potent and selective allosteric inhibitors of TYK2 signalling.
Medchemcomm, 8, 2017
|
|
5TCA
 
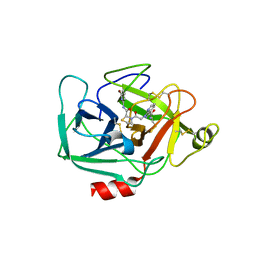 | | Complement Factor D inhibited with JH3 | | Descriptor: | 1-(2-{(2S)-2-[(6-bromopyridin-2-yl)carbamoyl]-1,3-thiazolidin-3-yl}-2-oxoethyl)-1H-pyrazolo[3,4-b]pyridine-3-carboxamide, Complement factor D | | Authors: | Stuckey, J.A. | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Buried Hydrogen Bond Interactions Contribute to the High Potency of Complement Factor D Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
5TKD
 
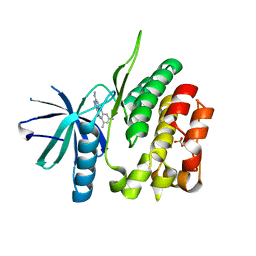 | | CRYSTAL STRUCTURE OF TYROSINE KINASE 2 JH2 (PSEUDO KINASE DOMAIN) COMPLEXED WITH 6-[(3,5-DIMETHYLPHE NYL)AMINO]-8- (METHYLAMINO)IMIDAZO[1,2-B]PYRIDAZINE-3-CARBO XAMIDE | | Descriptor: | 6-[(3,5-dimethylphenyl)amino]-8-(methylamino)imidazo[1,2-b]pyridazine-3-carboxamide, Non-receptor tyrosine-protein kinase TYK2, SULFATE ION | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2016-10-06 | | Release date: | 2016-12-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Identification of imidazo[1,2-b]pyridazine TYK2 pseudokinase ligands as potent and selective allosteric inhibitors of TYK2 signalling.
Medchemcomm, 8, 2017
|
|
5TCC
 
 | | Complement Factor D inhibited with JH4 | | Descriptor: | (2S)-N-(6-bromopyridin-2-yl)-3-[(1H-indazol-1-yl)acetyl]-1,3-thiazolidine-2-carboxamide, Complement factor D | | Authors: | Stuckey, J.A. | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Buried Hydrogen Bond Interactions Contribute to the High Potency of Complement Factor D Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
7YUY
 
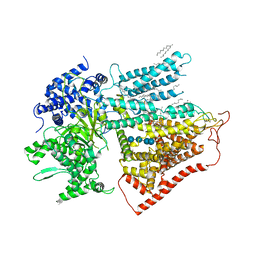 | | Structure of a mutated membrane-bound glycosyltransferase | | Descriptor: | (11R,14S)-17-amino-14-hydroxy-8,14-dioxo-9,13,15-trioxa-14lambda~5~-phosphaheptadecan-11-yl decanoate, 1,3-beta-glucan synthase component FKS1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, X.L, Yang, P, Zhang, M, Liu, X.T, Yu, H.J. | | Deposit date: | 2022-08-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and mechanistic insights into fungal beta-1,3-glucan synthase FKS1.
Nature, 616, 2023
|
|
7WWM
 
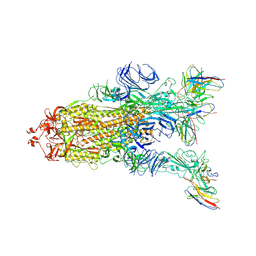 | | S protein of Delta variant in complex with ZWC6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-13 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Broadly neutralizing antibodies against Omicron-included SARS-CoV-2 variants induced by vaccination.
Signal Transduct Target Ther, 7, 2022
|
|
7VXG
 
 | |
7WWL
 
 | | S protein of Delta variant in complex with ZWD12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-13 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Broadly neutralizing antibodies against Omicron-included SARS-CoV-2 variants induced by vaccination.
Signal Transduct Target Ther, 7, 2022
|
|
7DIY
 
 | | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-exoribonuclease domain | | Descriptor: | MAGNESIUM ION, ZINC ION, nsp10 protein, ... | | Authors: | Lin, S, Chen, H, Chen, Z.M, Yang, F.L, Ye, F, Zheng, Y, Yang, J, Lin, X, Sun, H.L, Wang, L.L, Wen, A, Cao, Y, Lu, G.W. | | Deposit date: | 2020-11-19 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity.
Nucleic Acids Res., 49, 2021
|
|
5GM5
 
 | | Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellobiose | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endoglucanase-1, SULFATE ION, ... | | Authors: | Huang, J.W, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure and genetic modifications of FI-CMCase from Aspergillus aculeatus F-50
Biochem. Biophys. Res. Commun., 478, 2016
|
|
5H65
 
 | | Crystal structure of human POT1 and TPP1 | | Descriptor: | Adrenocortical dysplasia protein homolog, Protection of telomeres protein 1, ZINC ION | | Authors: | Chen, C, Wu, J, Lei, M. | | Deposit date: | 2016-11-10 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into POT1-TPP1 interaction and POT1 C-terminal mutations in human cancer.
Nat Commun, 8, 2017
|
|
6NZR
 
 | |
6NZE
 
 | |
6NZP
 
 | |
6OCQ
 
 | | Crystal structure of RIP1 kinase in complex with a pyrrolidine | | Descriptor: | 1-[(2S)-2-(3-fluorophenyl)pyrrolidin-1-yl]-2,2-dimethylpropan-1-one, Receptor-interacting serine/threonine-protein kinase 1, SULFATE ION | | Authors: | Thorpe, J.H, Harris, P.A. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Discovery and Lead-Optimization of 4,5-Dihydropyrazoles as Mono-Kinase Selective, Orally Bioavailable and Efficacious Inhibitors of Receptor Interacting Protein 1 (RIP1) Kinase.
J.Med.Chem., 62, 2019
|
|
6NZQ
 
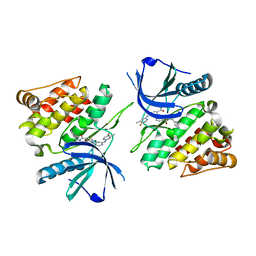 | |
6NZF
 
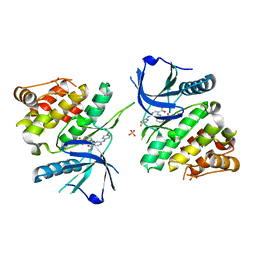 | |
6NZH
 
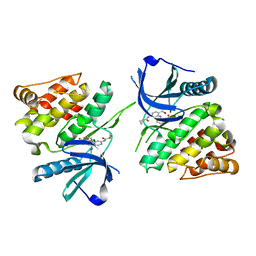 | |
5GM4
 
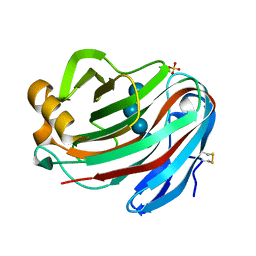 | | Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellotetrose | | Descriptor: | Endoglucanase-1, SULFATE ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Huang, J.W, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure and genetic modifications of FI-CMCase from Aspergillus aculeatus F-50
Biochem. Biophys. Res. Commun., 478, 2016
|
|
5GM3
 
 | | Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 | | Descriptor: | CACODYLATE ION, Endoglucanase-1, ZINC ION | | Authors: | Huang, J.W, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structure and genetic modifications of FI-CMCase from Aspergillus aculeatus F-50
Biochem. Biophys. Res. Commun., 478, 2016
|
|
5XXF
 
 | | Crystal structure of Poz1, Tpz1 and Rap1 | | Descriptor: | Protection of telomeres protein poz1, Protection of telomeres protein tpz1, Rap1, ... | | Authors: | Xue, J, Chen, H, Wu, J, Lei, M. | | Deposit date: | 2017-07-03 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the fission yeast S. pombe telomeric Tpz1-Poz1-Rap1 complex.
Cell Res., 27, 2017
|
|
