2OG8
 
 | | crystal structure of aminoquinazoline 36 bound to Lck | | Descriptor: | N-{2-[(N,N-DIETHYLGLYCYL)AMINO]-5-(TRIFLUOROMETHYL)PHENYL}-4-METHYL-3-[2-(METHYLAMINO)QUINAZOLIN-6-YL]BENZAMIDE, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2007-01-05 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Aminoquinazolines as Potent, Orally Bioavailable Inhibitors of Lck: Synthesis, SAR, and in Vivo Anti-Inflammatory Activity
J.Med.Chem., 49, 2006
|
|
2OO8
 
 | | Synthesis, Structural Analysis, and SAR Studies of Triazine Derivatives as Potent, Selective Tie-2 Inhibitors | | Descriptor: | Angiopoietin-1 receptor, N-{3-[3-(DIMETHYLAMINO)PROPYL]-5-(TRIFLUOROMETHYL)PHENYL}-4-METHYL-3-[(3-PYRIMIDIN-4-YLPYRIDIN-2-YL)AMINO]BENZAMIDE | | Authors: | Bellon, S.F, Kim, J. | | Deposit date: | 2007-01-25 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis, structural analysis, and SAR studies of triazine derivatives as potent, selective Tie-2 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2OSC
 
 | | Synthesis, Structural Analysis, and SAR Studies of Triazine Derivatives as Potent, Selective Tie-2 Inhibitors | | Descriptor: | Angiopoietin-1 receptor, N-{4-METHYL-3-[(3-PYRIMIDIN-4-YLPYRIDIN-2-YL)AMINO]PHENYL}-3-(TRIFLUOROMETHYL)BENZAMIDE | | Authors: | Bellon, S.F, Kim, J. | | Deposit date: | 2007-02-05 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synthesis, structural analysis, and SAR studies of triazine derivatives as potent, selective Tie-2 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2P4I
 
 | |
7YIR
 
 | |
7YIS
 
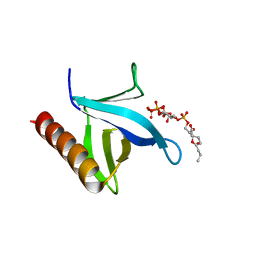 | | Crystal structure of N-terminal PH domain of ARAP3 protein in complex with inositol 1,3,4,5-tetrakisphosphate | | Descriptor: | (2R)-3-{[(S)-{[(2S,3R,5S,6S)-2,6-DIHYDROXY-3,4,5-TRIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-(1-HYDROXY BUTOXY)PROPYL BUTYRATE, Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3 | | Authors: | Zhang, Y.J, Liu, Y.R, Wu, B. | | Deposit date: | 2022-07-18 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Insights Uncover the Specific Phosphoinositide Recognition by the PH1 Domain of Arap3.
Int J Mol Sci, 24, 2023
|
|
4K4D
 
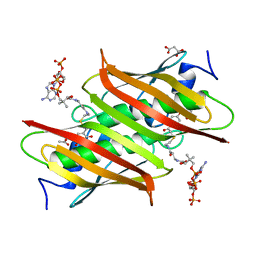 | | X-ray crystal structure of E. coli YbdB complexed with 2,4-dihydroxyphenacyl-CoA | | Descriptor: | 2,4-dihydroxyphenacyl coenzyme A, ACETATE ION, MALONATE ION, ... | | Authors: | Ru, W, Farelli, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2013-04-12 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and Catalysis in the Escherichia coli Hotdog-fold Thioesterase Paralogs YdiI and YbdB.
Biochemistry, 53, 2014
|
|
4K4C
 
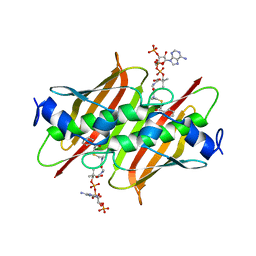 | | X-ray crystal structure of E. coli YbdB complexed with phenacyl-CoA | | Descriptor: | Proofreading thioesterase EntH, phenacyl coenzyme A | | Authors: | Ru, W, Farelli, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2013-04-12 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Catalysis in the Escherichia coli Hotdog-fold Thioesterase Paralogs YdiI and YbdB.
Biochemistry, 53, 2014
|
|
4K4B
 
 | | X-ray crystal structure of E. coli YdiI complexed with undeca-2-one-CoA | | Descriptor: | CHLORIDE ION, Esterase YdiI, undeca-2-one coenzyme A | | Authors: | Ru, W, Farelli, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2013-04-12 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Catalysis in the Escherichia coli Hotdog-fold Thioesterase Paralogs YdiI and YbdB.
Biochemistry, 53, 2014
|
|
4K4A
 
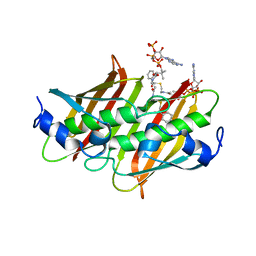 | | X-ray crystal structure of E. coli YdiI complexed with phenacyl-CoA | | Descriptor: | Esterase YdiI, phenacyl coenzyme A | | Authors: | Ru, W, Farelli, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2013-04-12 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and Catalysis in the Escherichia coli Hotdog-fold Thioesterase Paralogs YdiI and YbdB.
Biochemistry, 53, 2014
|
|
4K49
 
 | | X-ray crystal structure of E. coli YdiI complexed with 2,4-dihydroxyphenacyl CoA | | Descriptor: | 2,4-dihydroxyphenacyl coenzyme A, Esterase YdiI | | Authors: | Ru, W, Farelli, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2013-04-12 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and Catalysis in the Escherichia coli Hotdog-fold Thioesterase Paralogs YdiI and YbdB.
Biochemistry, 53, 2014
|
|
4JND
 
 | | Structure of a C.elegans sex determining protein | | Descriptor: | Ca(2+)/calmodulin-dependent protein kinase phosphatase, MAGNESIUM ION | | Authors: | Feng, Y, Zhang, Y, Ge, J, Yang, M. | | Deposit date: | 2013-03-15 | | Release date: | 2013-06-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Structural insight into Caenorhabditis elegans sex-determining protein FEM-2.
J.Biol.Chem., 288, 2013
|
|
4MKI
 
 | | Cobalt transporter ATP-binding subunit | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Energy-coupling factor transporter ATP-binding protein EcfA2, SULFATE ION | | Authors: | Yu, Y, Zhang, L, Chai, C.L, Heymann, D. | | Deposit date: | 2013-09-05 | | Release date: | 2013-10-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for a homodimeric ATPase subunit of an ECF transporter
Protein Cell, 4, 2013
|
|
8G92
 
 | | Structure of inhibitor 16d-bound SPNS2 | | Descriptor: | 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Chen, H, Li, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and functional insights into Spns2-mediated transport of sphingosine-1-phosphate.
Cell, 186, 2023
|
|
7X38
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 8A10 (CVB1-E:8A10) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3F
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 9A3 (CVB1-A:9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X42
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A-particle in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP0, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2G
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb nAb 2E6 (CVB1-E:2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-25 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2O
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 2E6 (CVB1-M:2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-25 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X49
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10 and 9A3) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2I
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A particle in complex with nAb 2E6 (CVB1-pre-A:2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-25 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2W
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A particle in complex with nAb 8A10 (CVB1-pre-A:8A10) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-26 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2T
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (CVB1-M:8A10) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-26 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X37
 
 | | Cryo-EM structure of Coxsackievirus B1 A particle in complex with nAb 2E6 (CVB1-A:2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3E
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A-particle in complex with nAb 9A3 (CVB1-pre-A:9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
