4CFU
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 3-[2-azanyl-6-(cyclohexylmethoxy)-7H-purin-8-yl]-2-methyl-benzoic acid, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2, ... | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-19 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
4CFV
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 3-[2-amino-6-(cyclohexylmethoxy)-7H-purin-8-yl]-2-methylphenol, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2, ... | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-19 | | Release date: | 2014-12-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
7Y36
 
 | | Cryo-EM structure of the Teriparatide-bound human PTH1R-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform Gnas-2 of Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Zhai, X, Mao, C, Shen, Q, Zang, S, Shen, D, Zhang, H, Chen, Z, Wang, G, Zhang, C, Zhang, Y, Liu, Z. | | Deposit date: | 2022-06-10 | | Release date: | 2023-07-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of the Teriparatide-bound human PTH1R-Gs complex
to be published
|
|
9FPD
 
 | | Crystal structure of human TAK1/TAB1 fusion protein in complex with compound S1 | | Descriptor: | 1,2-ETHANEDIOL, 6-[5-[6-(4-oxidanylcyclohexyl)oxy-1~{H}-pyrrolo[2,3-b]pyridin-5-yl]-1,2-oxazol-3-yl]pyridine-2-carboxamide, Mitogen-activated protein kinase kinase kinase 7,TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 | | Authors: | Scheufler, C, Lammens, A. | | Deposit date: | 2024-06-13 | | Release date: | 2024-12-04 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Identification of TAK-756, A Potent TAK1 Inhibitor for the Treatment of Osteoarthritis through Intra-Articular Administration.
J.Med.Chem., 67, 2024
|
|
4BTT
 
 | | factor Xa in complex with the dual thrombin-FXa inhibitor 31. | | Descriptor: | CALCIUM ION, COAGULATION FACTOR X, COAGULATION FACTOR X LIGHT CHAIN, ... | | Authors: | Meneyrol, J, Follmann, M, Lassalle, G, Wehner, V, Barre, G, Rousseaux, T, Altenburger, J.M, Petit, F, Bocskei, Z, Stehlin-Gaon, C, Schreuder, H, Alet, N, Herault, J.-P, Millet, L, Dol, F, Hasbrand, C, Schaeffer, P, Sadoun, F, Klieber, S, Briot, C, Bono, F, Herbert, J.-M. | | Deposit date: | 2013-06-19 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | 5-Chlorothiophene-2-Carboxylic Acid [(S)-2-[2-Methyl-3-(2-Oxopyrrolidin-1-Yl)Benzenesulfonylamino]-3-(4-Methylpiperazin-1-Yl)-3-Oxopropyl]Amide (Sar107375), a Selective and Potent Orally Active Dual Thrombin and Factor Xa Inhibitor.
J.Med.Chem., 56, 2013
|
|
4BTI
 
 | | factor Xa in complex with the dual thrombin-FXa inhibitor 58. | | Descriptor: | 5-Chloro-thiophene-2-carboxylic acid [(S)-2-[2-difluoromethoxy-3-(2-oxo-piperidin-1-yl)-benzenesulfonylamino]-3-((S)-3-dimethylamino-pyrrolidin-1-yl)-3-oxo-propyl]-amide, CALCIUM ION, COAGULATION FACTOR X HEAVY CHAIN, ... | | Authors: | Meneyrol, J, Follmann, M, Lassalle, G, Wehner, V, Barre, G, Rousseaux, T, Altenburger, J.M, Petit, F, Bocskei, Z, Stehlin-Gaon, C, Schreuder, H, Alet, N, Herault, J.-P, Millet, L, Dol, F, Hasbrand, C, Schaeffer, P, Sadoun, F, Klieber, S, Briot, C, Bono, F, Herbert, J.-M. | | Deposit date: | 2013-06-18 | | Release date: | 2013-12-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 5-Chlorothiophene-2-Carboxylic Acid [(S)-2-[2-Methyl-3-(2-Oxopyrrolidin-1-Yl)Benzenesulfonylamino]-3-(4-Methylpiperazin-1-Yl)-3-Oxopropyl]Amide (Sar107375), a Selective and Potent Orally Active Dual Thrombin and Factor Xa Inhibitor.
J.Med.Chem., 56, 2013
|
|
7BCQ
 
 | | ASCT2 in the presence of the inhibitor Lc-BPE (position "up") in the outward-open conformation. | | Descriptor: | 4-(4-phenylphenyl)carbonyloxypyrrolidine-2-carboxylic acid, Neutral amino acid transporter B(0) | | Authors: | Garibsingh, R.A, Ndaru, E, Garaeva, A.A, Shi, Y, Zielewicz, L, Bonomi, M, Slotboom, D.J, Paulino, C, Grewer, C, Schlessinger, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Rational design of ASCT2 inhibitors using an integrated experimental-computational approach.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6IAF
 
 | | Fifteen minutes iron loaded Rana Catesbeiana H' ferritin variant H54N | | Descriptor: | CHLORIDE ION, FE (II) ION, Ferritin, ... | | Authors: | Pozzi, C, Di Pisa, F, Mangani, S. | | Deposit date: | 2018-11-26 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Effect of the point mutation H54N on the ferroxidase process of Rana catesbeiana H' ferritin.
J.Inorg.Biochem., 197, 2019
|
|
7BCT
 
 | | ASCT2 in the presence of the inhibitor ERA-21 in the outward-open conformation. | | Descriptor: | Neutral amino acid transporter B(0) | | Authors: | Garibsingh, R.A, Ndaru, E, Garaeva, A.A, Shi, Y, Zielewicz, L, Bonomi, M, Slotboom, D.J, Paulino, C, Grewer, C, Schlessinger, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Rational design of ASCT2 inhibitors using an integrated experimental-computational approach.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4BQ7
 
 | | Crystal structure of the RGMB-Neo1 complex form 2 | | Descriptor: | NEOGENIN, RGM DOMAIN FAMILY MEMBER B | | Authors: | Bell, C.H, Healey, E, van Erp, S, Bishop, B, Tang, C, Gilbert, R.J.C, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | Deposit date: | 2013-05-30 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (6.601 Å) | | Cite: | Structure of the Repulsive Guidance Molecule (Rgm)-Neogenin Signaling Hub
Science, 341, 2013
|
|
6IC6
 
 | | Human cathepsin-C in complex with cyclopropyl peptidyl nitrile inhibitor 1 | | Descriptor: | (2~{S})-~{N}-[(1~{R},2~{R})-1-(aminomethyl)-2-[4-[4-(trifluoromethyl)phenyl]phenyl]cyclopropyl]-2-azanyl-butanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hakansson, M, Logan, D.T, Korkmaz, B, Lesner, A, Wysocka, M, Gieldon, A, Gauthier, F, Jenne, D, Lauritzen, C, Pedersen, J. | | Deposit date: | 2018-12-02 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Structure-based design and in vivo anti-arthritic activity evaluation of a potent dipeptidyl cyclopropyl nitrile inhibitor of cathepsin C.
Biochem. Pharmacol., 164, 2019
|
|
6IGB
 
 | | the structure of Pseudomonas aeruginosa Periplasmic gluconolactonase, PpgL | | Descriptor: | ACETATE ION, Periplasmic gluconolactonase, PpgL, ... | | Authors: | Song, Y.J, Shen, Y.L, Wang, K.L, Li, T, Zhu, Y.B, Li, C.C, He, L.H, Zhao, N.L, Zhao, C, Yang, J, Huang, Q, Mu, X.Y. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural and Functional Insights into PpgL, a Metal-Independent beta-Propeller Gluconolactonase That Contributes toPseudomonas aeruginosaVirulence.
Infect.Immun., 87, 2019
|
|
6ILX
 
 | | Crystal structure of PETase W159F mutant from Ideonella sakaiensis | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SODIUM ION | | Authors: | Liu, C.C, Shi, C. | | Deposit date: | 2018-10-20 | | Release date: | 2019-03-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional characterization of polyethylene terephthalate hydrolase from Ideonella sakaiensis.
Biochem. Biophys. Res. Commun., 508, 2019
|
|
5YHQ
 
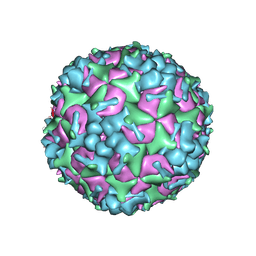 | | Cryo-EM Structure of CVA6 VLP | | Descriptor: | Capsid protein VP1, Capsid protein VP3, capsid protein VP0 | | Authors: | Chen, J, Zhang, C, Huang, Z, Cong, Y. | | Deposit date: | 2017-09-29 | | Release date: | 2017-10-25 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A 3.0-Angstrom Resolution Cryo-Electron Microscopy Structure and Antigenic Sites of Coxsackievirus A6-Like Particles.
J. Virol., 92, 2018
|
|
5YLY
 
 | |
5Y7L
 
 | |
5Z4Y
 
 | |
3CAO
 
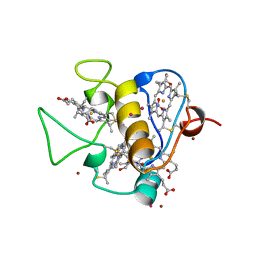 | | OXIDISED STRUCTURE OF THE ACIDIC CYTOCHROME C3 FROM DESULFOVIBRIO AFRICANUS | | Descriptor: | ARSENIC, CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Norager, S, Legrand, P, Pieulle, L, Hatchikian, C, Roth, M. | | Deposit date: | 1998-11-17 | | Release date: | 2000-07-23 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the oxidised and reduced acidic cytochrome c3from Desulfovibrio africanus.
J.Mol.Biol., 290, 1999
|
|
3CBT
 
 | | Crystal structure of SC4828, a unique phosphatase from Streptomyces coelicolor | | Descriptor: | MAGNESIUM ION, Phosphatase SC4828, SODIUM ION | | Authors: | Singer, A.U, Xu, X, Chang, C, Zheng, H, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-22 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of SC4828, a unique phosphatase from Streptomyces coelicolor.
To be Published
|
|
3CC9
 
 | | Crystal structure of Plasmodium vivax putative polyprenyl pyrophosphate synthase in complex with geranylgeranyl diphosphate | | Descriptor: | GERANYLGERANYL DIPHOSPHATE, Putative farnesyl pyrophosphate synthase, SODIUM ION | | Authors: | Wernimont, A.K, Dunford, J, Lew, J, Zhao, Y, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-25 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Plasmodium vivax putative polyprenyl pyrophosphate synthase in complex with geranylgeranyl diphosphate.
To be Published
|
|
3CAB
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera soaked at pH 7.0 | | Descriptor: | GLYCEROL, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-02-19 | | Release date: | 2008-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
3CBY
 
 | |
3CBX
 
 | |
3CDD
 
 | | Crystal structure of prophage MuSo2, 43 kDa tail protein from Shewanella oneidensis | | Descriptor: | Prophage MuSo2, 43 kDa tail protein | | Authors: | Chang, C, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-26 | | Release date: | 2008-03-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of prophage MuSo2, 43 kDa tail protein from Shewanella oneidensis.
To be Published
|
|
3CDU
 
 | | Crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase (3Dpol) in complex with a pyrophosphate | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gruez, A, Selisko, B, Roberts, M, Bricogne, G, Bussetta, C, Canard, B. | | Deposit date: | 2008-02-27 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases
J.Virol., 82, 2008
|
|
