4QO7
 
 | |
4R0I
 
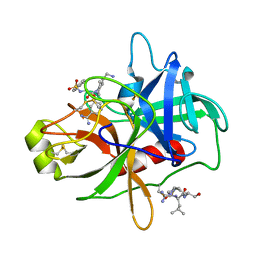 | | CRYSTAL STRUCTURE of MATRIPTASE in COMPLEX WITH INHIBITOR | | Descriptor: | 3-({(2S)-3-[4-(2-aminoethyl)piperidin-1-yl]-2-[(naphthalen-2-ylsulfonyl)amino]-3-oxopropyl}oxy)benzenecarboximidamide, SERINE PROTEASE, MATRIPTASE, ... | | Authors: | Rao, K.N, Ashok, K.N, Chakshusmathi, G, Rajeev, G, Subramanya, H. | | Deposit date: | 2014-07-31 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of O-(3-carbamimidoylphenyl)-l-serine amides as matriptase inhibitors using a fragment-linking approach
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4R68
 
 | | Lactate Dehydrogenase in complex with inhibitor compound 31 | | Descriptor: | (1S)-1-phenylethyl (4-chloro-3-{[(4S)-4-(2,6-dichlorophenyl)-2-hydroxy-6-oxocyclohex-1-en-1-yl]sulfanyl}phenyl)acetate, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2014-08-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Optimization of 5-(2,6-dichlorophenyl)-3-hydroxy-2-mercaptocyclohex-2-enones as potent inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 25, 2014
|
|
4ZVV
 
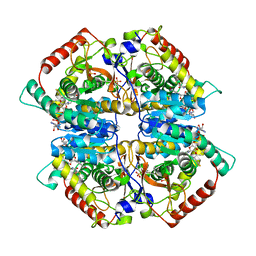 | | Lactate dehydrogenase A in complex with a trisubstituted piperidine-2,4-dione inhibitor GNE-140 | | Descriptor: | (2~{R})-5-(2-chlorophenyl)sulfanyl-2-(4-morpholin-4-ylphenyl)-4-oxidanyl-2-thiophen-3-yl-1,3-dihydropyridin-6-one, L-lactate dehydrogenase A chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Li, Y, Chen, Z, Eigenbrot, C. | | Deposit date: | 2015-05-18 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Metabolic plasticity underpins innate and acquired resistance to LDHA inhibition.
Nat.Chem.Biol., 12, 2016
|
|
5GUH
 
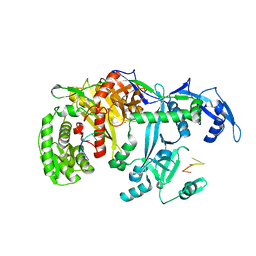 | | Crystal structure of silkworm PIWI-clade Argonaute Siwi bound to piRNA | | Descriptor: | MAGNESIUM ION, PIWI, RNA (28-MER) | | Authors: | Matsumoto, N, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Silkworm PIWI-Clade Argonaute Siwi Bound to piRNA
Cell, 167, 2016
|
|
2DYM
 
 | |
2DYO
 
 | |
4EI4
 
 | | JAK1 kinase (JH1 domain) in complex with compound 20 | | Descriptor: | (1R,3R)-3-(2-methylimidazo[4,5-d]pyrrolo[2,3-b]pyridin-1(8H)-yl)cyclohexanol, Tyrosine-protein kinase JAK1 | | Authors: | Eigenbrot, C, Steffek, M. | | Deposit date: | 2012-04-04 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery and optimization of C-2 methyl imidazopyrrolopyridines as potent and orally bioavailable JAK1 inhibitors with selectivity over JAK2.
J.Med.Chem., 55, 2012
|
|
4F08
 
 | |
4EHZ
 
 | | The Jak1 kinase domain in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-1-(piperidin-4-yl)-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Steffek, M. | | Deposit date: | 2012-04-04 | | Release date: | 2012-07-04 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Discovery and optimization of C-2 methyl imidazopyrrolopyridines as potent and orally bioavailable JAK1 inhibitors with selectivity over JAK2.
J.Med.Chem., 55, 2012
|
|
4FK6
 
 | | JAK1 kinase (JH1 domain) in complex with compound 72 | | Descriptor: | N-({1-[(1R,2R,4S)-bicyclo[2.2.1]hept-2-yl]-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridin-2-yl}methyl)methanesulfonamide, Tyrosine-protein kinase JAK1 | | Authors: | Eigenbrot, C, Steffek, M. | | Deposit date: | 2012-06-12 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based discovery of C-2 substituted imidazo-pyrrolopyridine JAK1 inhibitors with improved selectivity over JAK2.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4HGE
 
 | | JAK2 kinase (JH1 domain) in complex with compound 8 | | Descriptor: | N-[1-(3-chlorophenyl)-3-methyl-1H-pyrazol-5-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2012-10-08 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of potent and selective pyrazolopyrimidine janus kinase 2 inhibitors.
J.Med.Chem., 55, 2012
|
|
4F09
 
 | |
4HNJ
 
 | |
1EIQ
 
 | | 2,3-DIHYDROXYBIPHENYL-1,2-DIOXYGENASE | | Descriptor: | 2,3-DIHYDROXYBIPHENYL-1,2-DIOXYGENASE, FE (III) ION | | Authors: | Senda, T. | | Deposit date: | 2000-02-28 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of substrate free and complex forms of reactivated BphC, an extradiol type ring-cleavage dioxygenase.
J.Inorg.Biochem., 83, 2001
|
|
1YQY
 
 | | Structure of B. Anthrax Lethal factor in complex with a hydroxamate inhibitor | | Descriptor: | (2R)-2-{[(4-FLUORO-3-METHYLPHENYL)SULFONYL]AMINO}-N-HYDROXY-2-TETRAHYDRO-2H-PYRAN-4-YLACETAMIDE, Lethal factor, ZINC ION | | Authors: | Shoop, W.L, Xiong, Y, Wiltsie, J, Woods, A, Guo, J, Pivnichny, J.V, Felcetto, T, Michael, B.F, Bansal, A. | | Deposit date: | 2005-02-02 | | Release date: | 2005-05-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Anthrax lethal factor inhibition.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1GCS
 
 | | STRUCTURE OF THE BOVINE GAMMA-B CRYSTALLIN AT 150K | | Descriptor: | GAMMA-B CRYSTALLIN | | Authors: | Najmudin, S, Lindley, P, Slingsby, C, Bateman, O, Myles, D, Kumaraswamy, S, Glover, I. | | Deposit date: | 1994-01-27 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Bovine Gamma-B Crystallin at 150K
J.CHEM.SOC.,FARADAY TRANS., 89, 1993
|
|
1I5I
 
 | | THE C18S MUTANT OF BOVINE (GAMMA-B)-CRYSTALLIN | | Descriptor: | (GAMMA-B) CRYSTALLIN | | Authors: | Zarutskie, J.A, Asherie, N, Pande, J, Pande, A, Lomakin, J, Lomakin, A, Ogun, O, Stern, L.J, King, J.A, Benedek, G.B. | | Deposit date: | 2001-02-27 | | Release date: | 2001-03-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enhanced crystallization of the Cys18 to Ser mutant of bovine gammaB crystallin.
J.Mol.Biol., 314, 2001
|
|
2KAI
 
 | | REFINED 2.5 ANGSTROMS X-RAY CRYSTAL STRUCTURE OF THE COMPLEX FORMED BY PORCINE KALLIKREIN A AND THE BOVINE PANCREATIC TRYPSIN INHIBITOR. CRYSTALLIZATION, PATTERSON SEARCH, STRUCTURE DETERMINATION, REFINEMENT, STRUCTURE AND COMPARISON WITH ITS COMPONENTS AND WITH THE BOVINE TRYPSIN-PANCREATIC TRYPSIN INHIBITOR COMPLEX | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR, KALLIKREIN A | | Authors: | Bode, W, Chen, Z. | | Deposit date: | 1984-05-21 | | Release date: | 1984-07-19 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refined 2.5 A X-ray crystal structure of the complex formed by porcine kallikrein A and the bovine pancreatic trypsin inhibitor. Crystallization, Patterson search, structure determination, refinement, structure and comparison with its components and with the bovine trypsin-pancreatic trypsin inhibitor complex
J.Mol.Biol., 164, 1983
|
|
3SW8
 
 | |
3TQ7
 
 | | EB1c/EB3c heterodimer in complex with the CAP-Gly domain of P150glued | | Descriptor: | Dynactin subunit 1, Microtubule-associated protein RP/EB family member 1, Microtubule-associated protein RP/EB family member 3 | | Authors: | De Groot, C.O, Scharer, M.A, Capitani, G, Steinmetz, M.O. | | Deposit date: | 2011-09-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Interaction of mammalian end binding proteins with CAP-Gly domains of CLIP-170 and p150(glued).
J.Struct.Biol., 177, 2012
|
|
2TGP
 
 | | THE GEOMETRY OF THE REACTIVE SITE AND OF THE PEPTIDE GROUPS IN TRYPSIN, TRYPSINOGEN AND ITS COMPLEXES WITH INHIBITORS | | Descriptor: | CALCIUM ION, SULFATE ION, TRYPSIN INHIBITOR, ... | | Authors: | Huber, R, Bode, W, Deisenhofer, J, Schwager, P. | | Deposit date: | 1982-09-27 | | Release date: | 1983-01-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Geometry of the Reactive Site and of the Peptide Groups in Trypsin, Trypsinogen and its Complexes with Inhibitors
Acta Crystallogr.,Sect.B, 39, 1983
|
|
2LP2
 
 | | Solution structure and dynamics of human S100A1 protein modified at cysteine 85 with homocysteine disulfide bond formation in calcium saturated form | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, CALCIUM ION, Protein S100-A1 | | Authors: | Nowakowski, M.E, Jaremko, L, Jaremko, M, Zdanowski, K, Ejchart, A. | | Deposit date: | 2012-01-31 | | Release date: | 2013-02-20 | | Last modified: | 2024-04-03 | | Method: | SOLUTION NMR | | Cite: | Impact of calcium binding and thionylation of S100A1 protein on its nuclear magnetic resonance-derived structure and backbone dynamics.
Biochemistry, 52, 2013
|
|
2EAT
 
 | | Crystal structure of the SR CA2+-ATPASE with bound CPA and TG | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, OCTANOIC ACID [3S-[3ALPHA, 3ABETA, ... | | Authors: | Takahashi, M, Kondou, Y, Toyoshima, C. | | Deposit date: | 2007-02-02 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2EAU
 
 | | Crystal structure of the SR CA2+-ATPASE with bound CPA in the presence of curcumin | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, PHOSPHATIDYLETHANOLAMINE, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | | Authors: | Takahashi, M, Kondou, Y, Toyoshima, C. | | Deposit date: | 2007-02-02 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
