6KNV
 
 | | THRb mutation with a novel agonist | | Descriptor: | 2-[(1-methyl-4-oxidanyl-7-phenoxy-isoquinolin-3-yl)carbonylamino]ethanoic acid, Nuclear receptor coactivator 2, Thyroid hormone receptor beta | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2019-08-07 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Revealing a Mutant-Induced Receptor Allosteric Mechanism for the Thyroid Hormone Resistance.
Iscience, 20, 2019
|
|
6KKE
 
 | | THRb mutation with a novel agonist | | Descriptor: | 2-[(1-methyl-4-oxidanyl-7-phenoxy-isoquinolin-3-yl)carbonylamino]ethanoic acid, SRC2-3, Thyroid hormone receptor beta | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2019-07-25 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.577 Å) | | Cite: | Revealing a Mutant-Induced Receptor Allosteric Mechanism for the Thyroid Hormone Resistance.
Iscience, 20, 2019
|
|
6KKB
 
 | | A novel agonist of THRb | | Descriptor: | 2-[(1-methyl-4-oxidanyl-7-phenoxy-isoquinolin-3-yl)carbonylamino]ethanoic acid, SRC2-3, Thyroid hormone receptor beta | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2019-07-24 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Revealing a Mutant-Induced Receptor Allosteric Mechanism for the Thyroid Hormone Resistance.
Iscience, 20, 2019
|
|
6KNU
 
 | | THRb mutation with a novel agonist | | Descriptor: | 2-[(1-methyl-4-oxidanyl-7-phenoxy-isoquinolin-3-yl)carbonylamino]ethanoic acid, Nuclear receptor coactivator 2, Thyroid hormone receptor beta | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2019-08-07 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Revealing a Mutant-Induced Receptor Allosteric Mechanism for the Thyroid Hormone Resistance.
Iscience, 20, 2019
|
|
4WVD
 
 | | Identification of a novel FXR ligand that regulates metabolism | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, Bile acid receptor, FORMIC ACID, ... | | Authors: | Wang, R, Li, Y. | | Deposit date: | 2014-11-05 | | Release date: | 2015-02-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The antiparasitic drug ivermectin is a novel FXR ligand that regulates metabolism.
Nat Commun, 4, 2013
|
|
7XTO
 
 | |
6KTR
 
 | | Crystal structure of fibroblast growth factor 19 in complex with Fab | | Descriptor: | Fibroblast growth factor 19, G1A8-Fab-HC, G1A8-Fab-LC, ... | | Authors: | Liu, H, Zheng, S, Hou, X, Liu, X, Lv, X, Li, Y, Li, W, Sui, J. | | Deposit date: | 2019-08-28 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59775758 Å) | | Cite: | Novel Abs targeting the N-terminus of fibroblast growth factor 19 inhibit hepatocellular carcinoma growth without bile-acid-related side-effects.
Cancer Sci., 111, 2020
|
|
5JYY
 
 | | Structure-based Tetravalent Zanamivir with Potent Inhibitory Activity against Drug-resistant Influenza Viruses | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-[(2-methoxyethyl)carbamoyl]-D-glycero-D-galacto-non-2-enon ic acid, CALCIUM ION, ... | | Authors: | Fu, L, Wu, Y, Bi, Y, Zhang, S, Lv, X, Qi, J, Li, Y, Lu, X, Yan, J, Gao, G.F, Li, X. | | Deposit date: | 2016-05-15 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Tetravalent Zanamivir with Potent Inhibitory Activity against Drug-Resistant Influenza Viruses
J.Med.Chem., 59, 2016
|
|
5KO9
 
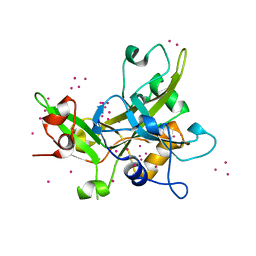 | | Crystal Structure of the SRAP Domain of Human HMCES Protein | | Descriptor: | Embryonic stem cell-specific 5-hydroxymethylcytosine-binding protein, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Li, Y, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-29 | | Release date: | 2016-08-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5MSZ
 
 | | Lytic Polysaccharide Monooxygenase AA15 from Thermobia domestica in the Cu(I) State | | Descriptor: | 1,2-ETHANEDIOL, COPPER (I) ION, Thermobia domestica domestica AA15 | | Authors: | Hemsworth, G.R, Sabbadin, F, Ciano, L, Henrissat, B, Dupree, P, Tryfona, T, Besser, K, Elias, L, Pesante, G, Li, Y, Dowle, A, Bates, R, Gomez, L, Hallam, R, Davies, G.J, Walton, P.H, Bruce, N.C, McQueen-Mason, S. | | Deposit date: | 2017-01-06 | | Release date: | 2018-02-28 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | An ancient family of lytic polysaccharide monooxygenases with roles in arthropod development and biomass digestion.
Nat Commun, 9, 2018
|
|
7YFG
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine and glutamate (major class in asymmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFH
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine, glutamate and (R)-PYD-106 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1C6V
 
 | | SIV INTEGRASE (CATALYTIC DOMAIN + DNA BIDING DOMAIN COMPRISING RESIDUES 50-293) MUTANT WITH PHE 185 REPLACED BY HIS (F185H) | | Descriptor: | PROTEIN (SIU89134), PROTEIN (SIV INTEGRASE) | | Authors: | Chen, Z, Yan, Y, Munshi, S, Li, Y, Zruygay-Murphy, J, Xu, B, Witmer, M, Felock, P, Wolfe, A, Sardana, V, Emini, E.A, Hazuda, D, Kuo, L.C. | | Deposit date: | 1999-12-21 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50-293)--an initial glance of the viral DNA binding platform.
J.Mol.Biol., 296, 2000
|
|
2AXH
 
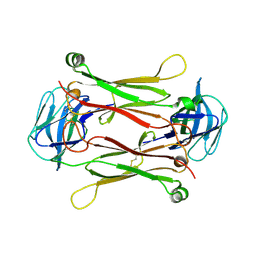 | | Crystal structures of T cell receptor beta chains related to rheumatoid arthritis | | Descriptor: | T cell receptor beta chain | | Authors: | Li, H, Van Vranken, S, Zhao, Y, Li, Z, Guo, Y, Eisele, L, Li, Y. | | Deposit date: | 2005-09-05 | | Release date: | 2005-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of T cell receptor (beta) chains related to rheumatoid arthritis.
Protein Sci., 14, 2005
|
|
2AXJ
 
 | | Crystal structures of T cell receptor beta chains related to rheumatoid arthritis | | Descriptor: | SF4 T cell receptor beta chain | | Authors: | Li, H, Van Vranken, S, Zhao, Y, Li, Z, Guo, Y, Eisele, L, Li, Y. | | Deposit date: | 2005-09-05 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of T cell receptor (beta) chains related to rheumatoid arthritis.
Protein Sci., 14, 2005
|
|
6VYC
 
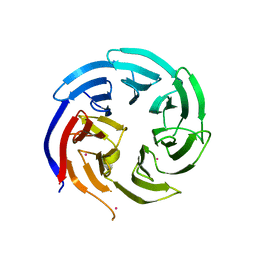 | | Crystal structure of WD-repeat domain of human WDR91 | | Descriptor: | UNKNOWN ATOM OR ION, WD repeat-containing protein 91 | | Authors: | Halabelian, L, Hutchinson, A, Li, Y, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a First-in-Class Small-Molecule Ligand for WDR91 Using DNA-Encoded Chemical Library Selection Followed by Machine Learning.
J.Med.Chem., 66, 2023
|
|
1Q57
 
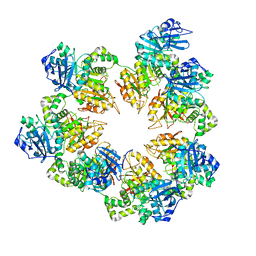 | | The Crystal Structure of the Bifunctional Primase-Helicase of Bacteriophage T7 | | Descriptor: | DNA primase/helicase | | Authors: | Toth, E.A, Li, Y, Sawaya, M.R, Cheng, Y, Ellenberger, T. | | Deposit date: | 2003-08-06 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The Crystal Structure of the Bifunctional Primase-Helicase of Bacteriophage T7
Mol.Cell, 12, 2003
|
|
6WSL
 
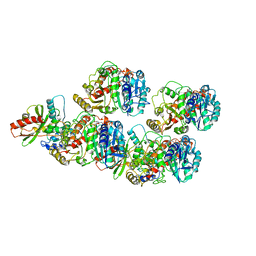 | | Cryo-EM structure of VASH1-SVBP bound to microtubules | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Small vasohibin-binding protein, ... | | Authors: | Li, F, Li, Y, Yu, H. | | Deposit date: | 2020-05-01 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of VASH1-SVBP bound to microtubules.
Elife, 9, 2020
|
|
1L4U
 
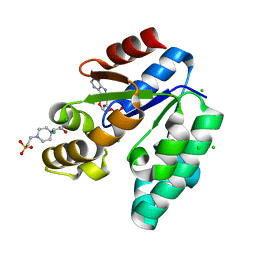 | | CRYSTAL STRUCTURE OF SHIKIMATE KINASE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MGADP AND PT(II) AT 1.8 ANGSTROM RESOLUTION | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Gu, Y, Reshetnikova, L, Li, Y, Wu, Y, Yan, H, Singh, S, Ji, X. | | Deposit date: | 2002-03-05 | | Release date: | 2002-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of shikimate kinase from Mycobacterium tuberculosis reveals the dynamic role of the LID domain in catalysis.
J.Mol.Biol., 319, 2002
|
|
1L4Y
 
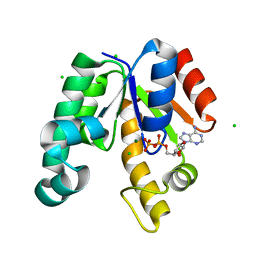 | | CRYSTAL STRUCTURE OF SHIKIMATE KINASE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MGADP AT 2.0 ANGSTROM RESOLUTION | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Gu, Y, Reshetnikova, L, Li, Y, Wu, Y, Yan, H, Singh, S, Ji, X. | | Deposit date: | 2002-03-06 | | Release date: | 2002-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of shikimate kinase from Mycobacterium tuberculosis reveals the dynamic role of the LID domain in catalysis.
J.Mol.Biol., 319, 2002
|
|
4X3G
 
 | | Crystal structure of SIAH1 SINA domain in complex with a USP19 peptide | | Descriptor: | E3 ubiquitin-protein ligase SIAH1, Ubiquitin carboxyl-terminal hydrolase 19, ZINC ION | | Authors: | Walker, J.R, Dong, A, Zhang, Q, Huang, X, Li, Y, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-11-28 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of SIAH1 SINA domain in complex with a USP19 peptide
To be published
|
|
1Z2E
 
 | | Solution Structure of Bacillus subtilis ArsC in oxidized state | | Descriptor: | Arsenate reductase | | Authors: | Jin, C, Li, Y. | | Deposit date: | 2005-03-08 | | Release date: | 2005-10-04 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structures and Backbone Dynamics of Arsenate Reductase from Bacillus subtilis: REVERSIBLE CONFORMATIONAL SWITCH ASSOCIATED WITH ARSENATE REDUCTION
J.Biol.Chem., 280, 2005
|
|
1BPH
 
 | | CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 | | Descriptor: | 1,2-DICHLOROETHANE, INSULIN A CHAIN (PH 9), INSULIN B CHAIN (PH 9), ... | | Authors: | Gursky, O, Badger, J, Li, Y, Caspar, D.L.D. | | Deposit date: | 1992-10-30 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes in cubic insulin crystals in the pH range 7-11.
Biophys.J., 63, 1992
|
|
4H53
 
 | | Influenza N2-Tyr406Asp neuraminidase in complex with beta-Neu5Ac | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Vavricka, C.J, Liu, Y, Kiyota, H, Sriwilaijaroen, N, Qi, J, Tanaka, K, Wu, Y, Li, Q, Li, Y, Yan, J, Suzuki, Y, Gao, G.F. | | Deposit date: | 2012-09-18 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Influenza neuraminidase operates via a nucleophilic mechanism and can be targeted by covalent inhibitors
Nat Commun, 4, 2013
|
|
1Z2D
 
 | | Solution Structure of Bacillus subtilis ArsC in reduced state | | Descriptor: | Arsenate reductase | | Authors: | Jin, C, Li, Y. | | Deposit date: | 2005-03-08 | | Release date: | 2005-10-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structures and Backbone Dynamics of Arsenate Reductase from Bacillus subtilis: REVERSIBLE CONFORMATIONAL SWITCH ASSOCIATED WITH ARSENATE REDUCTION
J.Biol.Chem., 280, 2005
|
|
