1AOZ
 
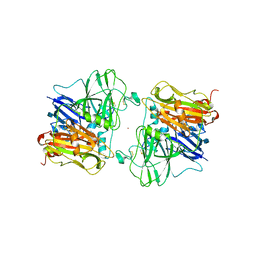 | | REFINED CRYSTAL STRUCTURE OF ASCORBATE OXIDASE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, COPPER (II) ION, ... | | Authors: | Messerschmidt, A, Ladenstein, R, Huber, R. | | Deposit date: | 1992-01-08 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Refined crystal structure of ascorbate oxidase at 1.9 A resolution.
J.Mol.Biol., 224, 1992
|
|
7U36
 
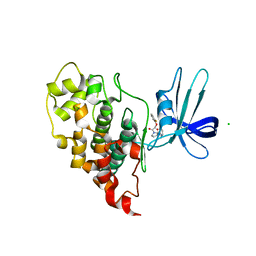 | | Crystal structure of human GSK3B in complex with ARN1484 | | Descriptor: | (3S)-1-[(2-fluorophenoxy)acetyl]-N-(pyridin-2-yl)pyrrolidine-3-carboxamide, CHLORIDE ION, Glycogen synthase kinase-3 beta | | Authors: | Tripathi, S.K, Balboni, B, Giabbai, B, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2022-02-25 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of Novel GSK-3 beta Hits Using Competitive Biophysical Assays.
Int J Mol Sci, 23, 2022
|
|
7U2Z
 
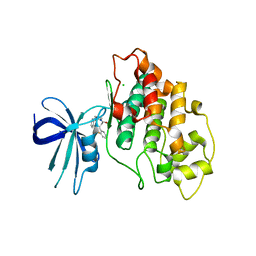 | | Crystal structure of human GSK3B in complex with G12 | | Descriptor: | (3R)-1-[3-(2-fluorophenyl)propanoyl]-N-(pyridin-2-yl)pyrrolidine-3-carboxamide, CHLORIDE ION, Glycogen synthase kinase-3 beta | | Authors: | Tripathi, S.K, Balboni, B, Giabbai, B, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2022-02-25 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Identification of Novel GSK-3 beta Hits Using Competitive Biophysical Assays.
Int J Mol Sci, 23, 2022
|
|
7U31
 
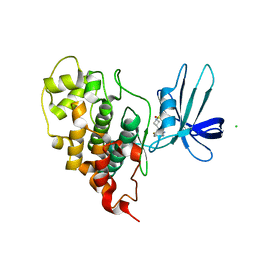 | | Crystal structure of human GSK3B in complex with G5 | | Descriptor: | 5-(4-fluorophenyl)-4-[1-(methanesulfonyl)azetidin-3-yl]pyrimidin-2-amine, CHLORIDE ION, Glycogen synthase kinase-3 beta | | Authors: | Tripathi, S.K, Balboni, B, Giabbai, B, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2022-02-25 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Identification of Novel GSK-3 beta Hits Using Competitive Biophysical Assays.
Int J Mol Sci, 23, 2022
|
|
7U33
 
 | | Crystal structure of human GSK3B in complex with ARN9133 | | Descriptor: | 3-[2-amino-5-(4-fluorophenyl)pyrimidin-4-yl]-N,N-dimethylazetidine-1-sulfonamide, CHLORIDE ION, Glycogen synthase kinase-3 beta | | Authors: | Tripathi, S.K, Balboni, B, Giabbai, B, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2022-02-25 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Novel GSK-3 beta Hits Using Competitive Biophysical Assays.
Int J Mol Sci, 23, 2022
|
|
6RCZ
 
 | | The structure of Burkholderia pseudomallei trehalose-6-phosphatase | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Trehalose 6-phosphate phosphatase | | Authors: | Gourlay, L.J. | | Deposit date: | 2019-04-12 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Functional and structural analysis of trehalose-6-phosphate phosphatase from Burkholderia pseudomallei: Insights into the catalytic mechanism.
Biochem.Biophys.Res.Commun., 523, 2020
|
|
6RJD
 
 | | Cryo-EM structure of St1Cas9-sgRNA-tDNA59-ntPAM complex. | | Descriptor: | Streptococcus Thermophilus 1 Cas9, ntPAM, sgRNA (78-MER), ... | | Authors: | Goulet, A, Chaves-Sanjuan, A, Cambillau, C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6.
Mol.Cell, 76, 2019
|
|
6RJ9
 
 | | Cryo-EM structure of St1Cas9-sgRNA-tDNA20-AcrIIA6 monomeric assembly. | | Descriptor: | AcrIIA6, CRISPR-associated endonuclease Cas9 1, sgRNA, ... | | Authors: | Goulet, A, Chaves-Sanjuan, A, Cambillau, C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6.
Mol.Cell, 76, 2019
|
|
6RJA
 
 | | Cryo-EM structure of St1Cas9-sgRNA-tDNA20-AcrIIA6 dimeric assembly. | | Descriptor: | AcrIIA6, CRISPR-associated endonuclease Cas9 1, RNA (78-MER), ... | | Authors: | Goulet, A, Cambillau, C, Chaves-Sanjuan, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6.
Mol.Cell, 76, 2019
|
|
6RJG
 
 | | Cryo-EM structure of St1Cas9-sgRNA-AcrIIA6-tDNA59-ntPAM complex. | | Descriptor: | AcrIIA6, Cas 9, ntPAM, ... | | Authors: | Goulet, A, Chaves-Sanjuan, A, Cambillau, C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6.
Mol.Cell, 76, 2019
|
|
5O7G
 
 | |
8CPE
 
 | |
7OFN
 
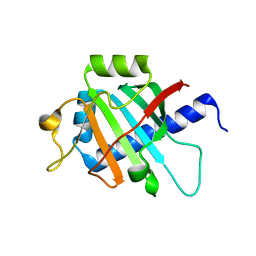 | |
5M9F
 
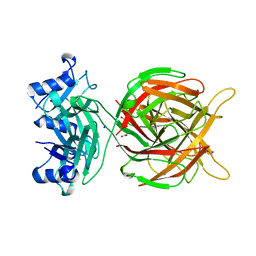 | |
1ASP
 
 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, COPPER (II) ION, ... | | Authors: | Messerschmidt, A, Luecke, H, Huber, R. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
6G4H
 
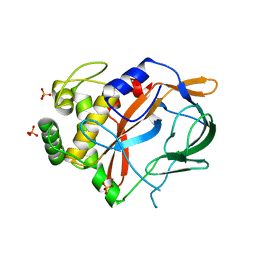 | | Crystal structure of the periplasmic domain of TgpA from Pseudomonas aeruginosa bound to ethylmercury | | Descriptor: | ETHYL MERCURY ION, PHOSPHATE ION, Protein-glutamine gamma-glutamyltransferase | | Authors: | Milani, M, Mastrangelo, E, Uruburu, M. | | Deposit date: | 2018-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional characterization of TgpA, a critical protein for the viability of Pseudomonas aeruginosa.
J.Struct.Biol., 205, 2019
|
|
6G49
 
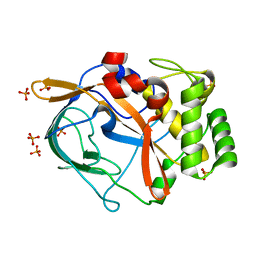 | | Crystal structure of the periplasmic domain of TgpA from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Protein-glutamine gamma-glutamyltransferase | | Authors: | Milani, M, Mastrangelo, E, Uruburu, M. | | Deposit date: | 2018-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional characterization of TgpA, a critical protein for the viability of Pseudomonas aeruginosa.
J.Struct.Biol., 205, 2019
|
|
6HUD
 
 | |
3MUP
 
 | | cIAP1-BIR3 domain in complex with the Smac-mimetic compound Smac037 | | Descriptor: | (3S,6S,7R,9aS)-6-{[(2S)-2-aminobutanoyl]amino}-7-(2-aminoethyl)-N-(diphenylmethyl)-5-oxooctahydro-1H-pyrrolo[1,2-a]azepine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Cossu, F, Malvezzi, F, Canevari, G, Milani, M. | | Deposit date: | 2010-05-03 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition of Smac-mimetic compounds by the BIR domain of cIAP1
Protein Sci., 19, 2010
|
|
1ASO
 
 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, COPPER (II) ION, ... | | Authors: | Messerschmidt, A, Luecke, H, Huber, R. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
1ASQ
 
 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, AZIDE ION, ... | | Authors: | Messerschmidt, A, Luecke, H, Huber, R. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
5FAF
 
 | | N184K pathological variant of gelsolin domain 2 (orthorhombic form) | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Boni, F, Milani, M, Ricagno, s, Bolognesi, M, de Rosa, M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Molecular basis of a novel renal amyloidosis due to N184K gelsolin variant.
Sci Rep, 6, 2016
|
|
8Q7D
 
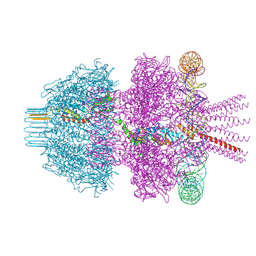 | | Neck of phage 812 after tail contraction (C12) | | Descriptor: | DNA forward strand (120-MER), DNA reverse strand (120-MER), Portal protein, ... | | Authors: | Cienikova, Z, Siborova, M, Fuzik, T, Plevka, P. | | Deposit date: | 2023-08-16 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Genome anchoring, retention, and release by neck proteins of Herelleviridae phage 812
To Be Published
|
|
8QEM
 
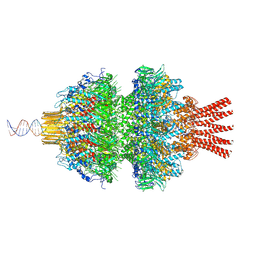 | | Neck channel of phage 812 after tail contraction (C1) | | Descriptor: | Channel DNA forward strand (63-MER), Channel DNA reverse strand (63-MER), Portal protein, ... | | Authors: | Cienikova, Z, Siborova, M, Fuzik, T, Plevka, P. | | Deposit date: | 2023-08-31 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Genome anchoring, retention, and release by neck proteins of Herelleviridae phage 812
To Be Published
|
|
8QEK
 
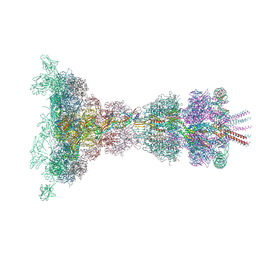 | | Neck and tail of phage 812 after tail contraction (composite) | | Descriptor: | Anchor DNA forward strand (120-MER), Anchor DNA reverse strand (120-MER), Baseplate hub assembly protein, ... | | Authors: | Cienikova, Z, Siborova, M, Fuzik, T, Plevka, P. | | Deposit date: | 2023-08-31 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Genome anchoring, retention, and release by neck proteins of Herelleviridae phage 812
To Be Published
|
|
