7EM3
 
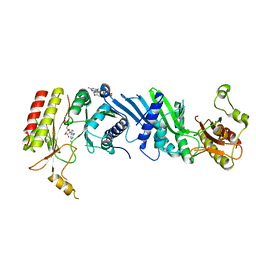 | | Crystal structure of the PI5P4Kbeta-2a-ATP complex | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta, [(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(azanyl)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphono hydrogen phosphate, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(azanyl)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2021-04-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 30, 2022
|
|
7EM4
 
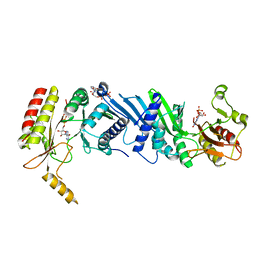 | | Crystal structure of the PI5P4Kbeta F205L-ITP complex | | Descriptor: | INOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta, [[(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-(6-oxidanylidene-1~{H}-purin-9-yl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2021-04-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 30, 2022
|
|
7R0L
 
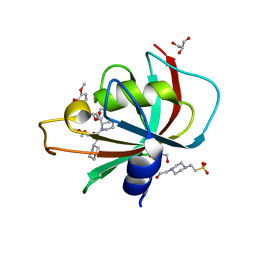 | | Structure of the FK1 domain of the FKBP51 G64S variant in complex with SAFit1 | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Meyners, C, Hausch, F. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Binding pocket stabilization by high-throughput screening of yeast display libraries.
Front Mol Biosci, 9, 2022
|
|
6VBX
 
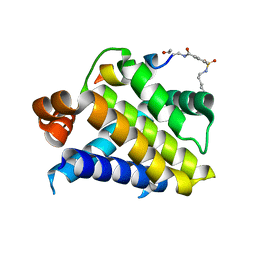 | | Crystal structure of Mcl-1 in complex with 138E12 peptide, Lys-covalent antagonist | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, Synthetic peptide | | Authors: | Pellecchia, M, Perry, J.J, Kenjic, N, Assar, Z. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, Synthesis, and Structural Characterization of Lysine Covalent BH3 Peptides Targeting Mcl-1.
J.Med.Chem., 64, 2021
|
|
5AXQ
 
 | | Crystal structure of the catalytic domain of PDE10A complexed with highly potent and brain-penetrant PDE10A Inhibitor with 2-oxindole scaffold | | Descriptor: | 1-(cyclopropylmethyl)-4-fluoranyl-5-[5-methoxy-4-oxidanylidene-3-(2-phenylpyrazol-3-yl)pyridazin-1-yl]-3,3-dimethyl-indol-2-one, 3-[3-fluoranyl-4-[5-methoxy-4-oxidanylidene-3-(2-phenylpyrazol-3-yl)pyridazin-1-yl]phenyl]-1,3-oxazolidin-2-one, MAGNESIUM ION, ... | | Authors: | Oki, H, Zama, Y. | | Deposit date: | 2015-07-31 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Design and synthesis of a novel 2-oxindole scaffold as a highly potent and brain-penetrant phosphodiesterase 10A inhibitor
Bioorg.Med.Chem., 23, 2015
|
|
1GH8
 
 | |
6UK2
 
 | |
6UJO
 
 | |
6UK4
 
 | |
6UJQ
 
 | |
5NWE
 
 | | Complex of H275Y mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
5NZF
 
 | | Complex of H275Y/I223V mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2017-05-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
5NZ4
 
 | | Complex of I223V mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J, Karlukova, E. | | Deposit date: | 2017-05-12 | | Release date: | 2018-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
5NZN
 
 | | Complex of H275Y/S247N mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2017-05-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
5NZE
 
 | | Complex of S247N mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J, Hejdanek, J. | | Deposit date: | 2017-05-13 | | Release date: | 2018-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
1GH9
 
 | |
6DGF
 
 | | Ubiquitin Variant bound to USP2 | | Descriptor: | Polyubiquitin-B, SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 2, ... | | Authors: | Manczyk, N, Sicheri, F. | | Deposit date: | 2018-05-17 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Yeast Two-Hybrid Analysis for Ubiquitin Variant Inhibitors of Human Deubiquitinases.
J. Mol. Biol., 431, 2019
|
|
5TTZ
 
 | |
6K4H
 
 | | Crystal structure of the PI5P4Kbeta-AMPPNP complex | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Senda, M, Senda, T. | | Deposit date: | 2019-05-23 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 2022
|
|
6HP0
 
 | | Complex of Neuraminidase from H1N1 Influenza Virus in Complex with Oseltamivir Triazol Derivative | | Descriptor: | (3~{R},4~{R},5~{S})-4-acetamido-5-[4-(hydroxymethyl)-1,2,3-triazol-1-yl]-3-pentan-3-yloxy-cyclohexene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2018-09-19 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Investigation of flexibility of neuraminidase 150-loop using tamiflu derivatives in influenza A viruses H1N1 and H5N1.
Bioorg.Med.Chem., 27, 2019
|
|
6K4G
 
 | | Crystal structure of the PI5P4Kbeta-GMPPNP complex | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Senda, M, Senda, T. | | Deposit date: | 2019-05-23 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 2022
|
|
4PGO
 
 | | Crystal structure of hypothetical protein PF0907 from Pyrococcus furiosus solved by sulfur SAD using Swiss Light Source data | | Descriptor: | CHLORIDE ION, Uncharacterized protein | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-05-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4PII
 
 | | Crystal structure of hypothetical protein PF0907 from pyrococcus furiosus solved by sulfur SAD using Swiss light source data | | Descriptor: | CHLORIDE ION, IMIDAZOLE, N-glycosylase/DNA lyase | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-05-08 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
1HYB
 
 | | CRYSTAL STRUCTURE OF AN ACTIVE SITE MUTANT OF METHANOBACTERIUM THERMOAUTOTROPHICUM NICOTINAMIDE MONONUCLEOTIDE ADENYLYLTRANSFERASE | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NICOTINAMIDE MONONUCLEOTIDE ADENYLYLTRANSFERASE, SULFATE ION | | Authors: | Saridakis, V, Christendat, D, Kimber, M.S, Edwards, A.M, Pai, E.F. | | Deposit date: | 2001-01-18 | | Release date: | 2001-03-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into ligand binding and catalysis of a central step in NAD+ synthesis: structures of Methanobacterium thermoautotrophicum NMN adenylyltransferase complexes.
J.Biol.Chem., 276, 2001
|
|
1QB4
 
 | | CRYSTAL STRUCTURE OF MN(2+)-BOUND PHOSPHOENOLPYRUVATE CARBOXYLASE | | Descriptor: | ASPARTIC ACID, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE CARBOXYLASE | | Authors: | Matsumura, H, Terada, M, Shirakata, S, Inoue, T, Yoshinaga, T, Izui, K, Kai, Y. | | Deposit date: | 1999-04-30 | | Release date: | 2002-05-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Plausible phosphoenolpyruvate binding site revealed by 2.6 A structure of Mn2+-bound phosphoenolpyruvate carboxylase from Escherichia coli
FEBS Lett., 458, 1999
|
|
