3WAV
 
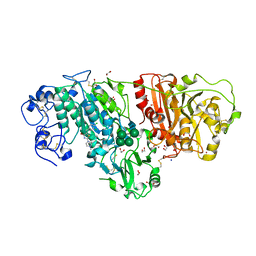 | | Crystal Structure of Autotaxin in Complex with Compound 10 | | Descriptor: | (5Z)-5-(3,4-dichlorobenzylidene)-2-(4-methylpiperazin-1-yl)-1,3-thiazol-4(5H)-one, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-09 | | Release date: | 2013-07-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Screening and X-ray Crystal Structure-based Optimization of Autotaxin (ENPP2) Inhibitors, Using a Newly Developed Fluorescence Probe
Acs Chem.Biol., 8, 2013
|
|
1UFA
 
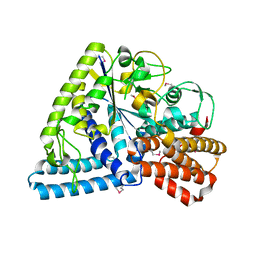 | | Crystal structure of TT1467 from Thermus thermophilus HB8 | | Descriptor: | TT1467 protein | | Authors: | Idaka, M, Terada, T, Murayama, K, Yamaguchi, H, Nureki, O, Ishitani, R, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-28 | | Release date: | 2003-11-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of TT1467 from Thermus thermophilus HB8
To be published
|
|
1V33
 
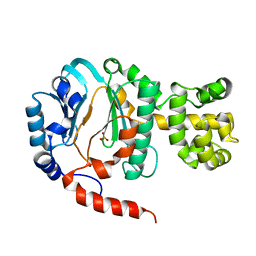 | | Crystal structure of DNA primase from Pyrococcus horikoshii | | Descriptor: | DNA primase small subunit, PHOSPHATE ION, ZINC ION | | Authors: | Ito, N, Nureki, O, Shirouzu, M, Yokoyama, S, Hanaoka, F, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-25 | | Release date: | 2004-03-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Pyrococcus horikoshii DNA primase-UTP complex: implications for the mechanism of primer synthesis.
Genes Cells, 8, 2003
|
|
1UDS
 
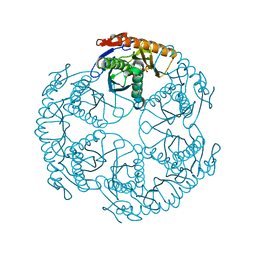 | | Crystal structure of the tRNA processing enzyme RNase PH R126A mutant from Aquifex aeolicus | | Descriptor: | PHOSPHATE ION, Ribonuclease PH, SULFATE ION | | Authors: | Ishii, R, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-02 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the tRNA Processing Enzyme RNase PH from Aquifex aeolicus
J.Biol.Chem., 278, 2003
|
|
3X3B
 
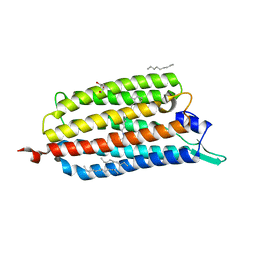 | | Crystal structure of the light-driven sodium pump KR2 in acidic state | | Descriptor: | DI(HYDROXYETHYL)ETHER, OLEIC ACID, RETINAL, ... | | Authors: | Kato, H.E, Inoue, K, Abe-Yoshizumi, R, Kato, Y, Ono, H, Konno, M, Ishizuka, T, Hoque, M.R, Hososhima, S, Kunitomo, H, Ito, J, Yoshizawa, S, Yamashita, K, Takemoto, M, Nishizawa, T, Taniguchi, R, Kogure, K, Maturana, A.D, Iino, Y, Yawo, H, Ishitani, R, Kandori, H, Nureki, O. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for Na(+) transport mechanism by a light-driven Na(+) pump
Nature, 521, 2015
|
|
1VFG
 
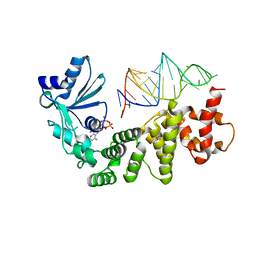 | | Crystal structure of tRNA nucleotidyltransferase complexed with a primer tRNA and an incoming ATP analog | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, RNA (75-MER), poly A polymerase | | Authors: | Tomita, K, Fukai, S, Ishitani, R, Ueda, T, Takeuchi, N, Vassylyev, D.G, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-04-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for template-independent RNA polymerization.
Nature, 430, 2004
|
|
1V34
 
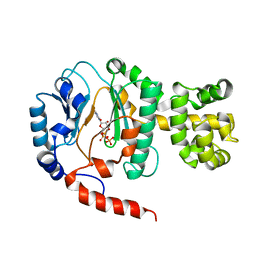 | | Crystal structure of Pyrococcus horikoshii DNA primase-UTP complex | | Descriptor: | DNA primase small subunit, URIDINE 5'-TRIPHOSPHATE, ZINC ION | | Authors: | Ito, N, Nureki, O, Shirouzu, M, Yokoyama, S, Hanaoka, F, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-25 | | Release date: | 2004-03-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Pyrococcus horikoshii DNA primase-UTP complex: implications for the mechanism of primer synthesis.
Genes Cells, 8, 2003
|
|
3X3C
 
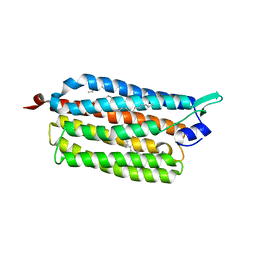 | | Crystal structure of the light-driven sodium pump KR2 in neutral state | | Descriptor: | OLEIC ACID, RETINAL, Sodium pumping rhodopsin | | Authors: | Kato, H.E, Inoue, K, Abe-Yoshizumi, R, Kato, Y, Ono, H, Konno, M, Ishizuka, T, Hoque, M.R, Hososhima, S, Kunitomo, H, Ito, J, Yoshizawa, S, Yamashita, K, Takemoto, M, Nishizawa, T, Taniguchi, R, Kogure, K, Maturana, A.D, Iino, Y, Yawo, H, Ishitani, R, Kandori, H, Nureki, O. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for Na(+) transport mechanism by a light-driven Na(+) pump
Nature, 521, 2015
|
|
1ULH
 
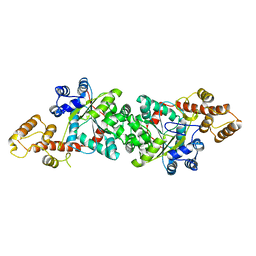 | | A short peptide insertion crucial for angiostatic activity of human tryptophanyl-tRNA synthetase | | Descriptor: | Tryptophanyl-tRNA synthetase | | Authors: | Kise, Y, Sengoku, T, Ishii, R, Yokoyama, S, Park, S.G, Lee, S.W, Kim, S, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-12 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A short peptide insertion crucial for angiostatic activity of human tryptophanyl-tRNA synthetase
Nat.Struct.Mol.Biol., 11, 2004
|
|
5X2G
 
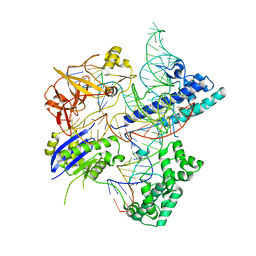 | | Crystal structure of Campylobacter jejuni Cas9 in complex with sgRNA and target DNA (AGAAACC PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, Non-target DNA strand, ... | | Authors: | Yamada, M, Watanabe, Y, Hirano, H, Nakane, T, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems
Mol. Cell, 65, 2017
|
|
5X2H
 
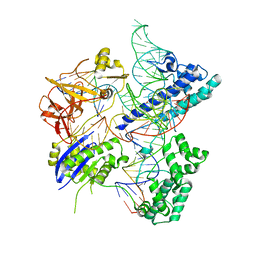 | | Crystal structure of Campylobacter jejuni Cas9 in complex with sgRNA and target DNA (AGAAACA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, Non-target DNA strand, ... | | Authors: | Yamada, M, Watanabe, Y, Hirano, H, Nakane, T, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems
Mol. Cell, 65, 2017
|
|
5WZY
 
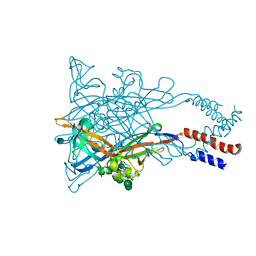 | | Crystal structure of the P2X4 receptor from zebrafish in the presence of CTP at 2.8 Angstroms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Kasuya, G, Hattori, M, Nureki, O. | | Deposit date: | 2017-01-19 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structural insights into the nucleotide base specificity of P2X receptors
Sci Rep, 7, 2017
|
|
5X93
 
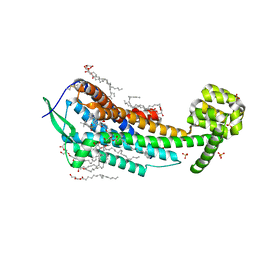 | | Human endothelin receptor type-B in complex with antagonist K-8794 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[6-[(4-tert-butylphenyl)sulfonylamino]-5-(2-methoxyphenoxy)-2-pyrimidin-2-yl-pyrimidin-4-yl]oxy-N-(2,6-dimethylphenyl)propanamide, CHOLESTEROL, ... | | Authors: | Shihoya, W, Nishizawa, T, Yamashita, K, Hirata, K, Okuta, A, Tani, K, Fujiyoshi, Y, Doi, T, Nureki, O. | | Deposit date: | 2017-03-05 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures of endothelin ETB receptor bound to clinical antagonist bosentan and its analog
Nat. Struct. Mol. Biol., 24, 2017
|
|
5X9H
 
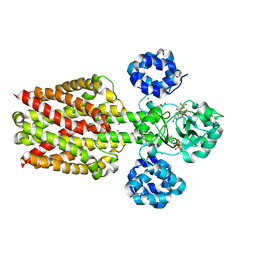 | |
5X9G
 
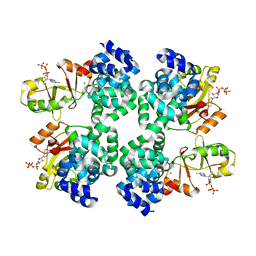 | |
5XPR
 
 | | Human endothelin receptor type-B in complex with antagonist bosentan | | Descriptor: | 4-tert-butyl-N-[6-(2-hydroxyethyloxy)-5-(2-methoxyphenoxy)-2-pyrimidin-2-yl-pyrimidin-4-yl]benzenesulfonamide, Endothelin B receptor,Endolysin,Endothelin B receptor, SULFATE ION | | Authors: | Shihoya, W, Nishizawa, T, Yamashita, K, Hirata, K, Okuta, A, Tani, K, Fujiyoshi, Y, Doi, T, Nureki, O. | | Deposit date: | 2017-06-04 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | X-ray structures of endothelin ETB receptor bound to clinical antagonist bosentan and its analog
Nat. Struct. Mol. Biol., 24, 2017
|
|
5XSZ
 
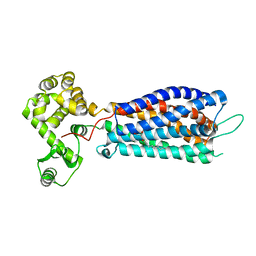 | | Crystal structure of zebrafish lysophosphatidic acid receptor LPA6 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lysophosphatidic acid receptor 6a,Endolysin,Lysophosphatidic acid receptor 6a | | Authors: | Taniguchi, R, Nishizawa, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-16 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into ligand recognition by the lysophosphatidic acid receptor LPA6
Nature, 548, 2017
|
|
5XH7
 
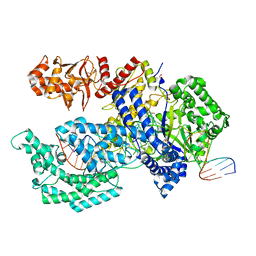 | | Crystal structure of the Acidaminococcus sp. BV3L6 Cpf1 RR variant in complex with crRNA and target DNA (TCCA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CRISPR-associated endonuclease Cpf1, ... | | Authors: | Nishimasu, H, Yamano, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Altered PAM Recognition by Engineered CRISPR-Cpf1
Mol. Cell, 67, 2017
|
|
5XUZ
 
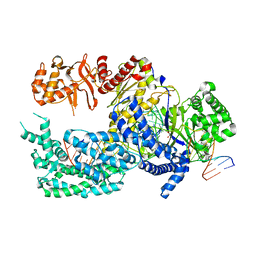 | | Crystal structure of Lachnospiraceae bacterium ND2006 Cpf1 in complex with crRNA and target DNA (CCCA PAM) | | Descriptor: | 1,2-ETHANEDIOL, DNA (29-MER), DNA (5'-D(*CP*GP*TP*CP*CP*CP*CP*CP*A)-3'), ... | | Authors: | Yamano, T, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-26 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Mol. Cell, 67, 2017
|
|
5XUT
 
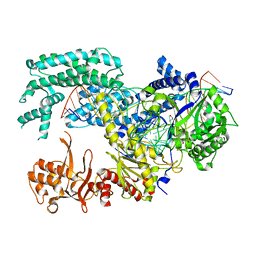 | | Crystal structure of Lachnospiraceae bacterium ND2006 Cpf1 in complex with crRNA and target DNA (TCTA PAM) | | Descriptor: | 1,2-ETHANEDIOL, DNA (29-MER), DNA (5'-D(*CP*GP*TP*CP*CP*TP*CP*TP*A)-3'), ... | | Authors: | Yamano, T, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-26 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Mol. Cell, 67, 2017
|
|
5XUS
 
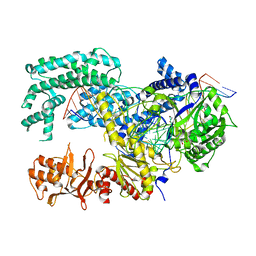 | | Crystal structure of Lachnospiraceae bacterium ND2006 Cpf1 in complex with crRNA and target DNA (TTTA PAM) | | Descriptor: | 1,2-ETHANEDIOL, DNA (29-MER), DNA (5'-D(*CP*GP*TP*CP*CP*TP*TP*TP*A)-3'), ... | | Authors: | Yamano, T, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-26 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Mol. Cell, 67, 2017
|
|
5XH6
 
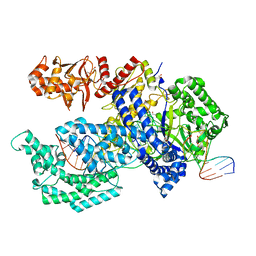 | | Crystal structure of the Acidaminococcus sp. BV3L6 Cpf1 RVR variant in complex with crRNA and target DNA (TATA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CRISPR-associated endonuclease Cpf1, ... | | Authors: | Nishimasu, H, Yamano, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Altered PAM Recognition by Engineered CRISPR-Cpf1
Mol. Cell, 67, 2017
|
|
5XUU
 
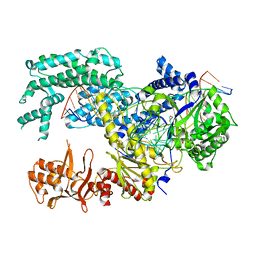 | | Crystal structure of Lachnospiraceae bacterium ND2006 Cpf1 in complex with crRNA and target DNA (TCCA PAM) | | Descriptor: | 1,2-ETHANEDIOL, DNA (29-MER), DNA (5'-D(*CP*GP*TP*CP*CP*TP*CP*CP*A)-3'), ... | | Authors: | Yamano, T, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-26 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Mol. Cell, 67, 2017
|
|
4X5M
 
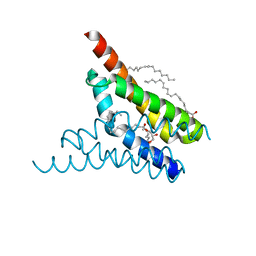 | | Crystal structure of SemiSWEET in the inward-open conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, OLEIC ACID, ... | | Authors: | Lee, Y, Nishizawa, T, Yamashita, K, Ishitani, R, Nureki, O. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the facilitative diffusion mechanism by SemiSWEET transporter
Nat Commun, 6, 2015
|
|
4X5N
 
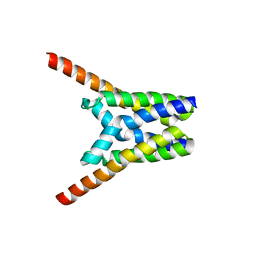 | | Crystal structure of SemiSWEET in the inward-open and outward-open conformations | | Descriptor: | Uncharacterized protein | | Authors: | Lee, Y, Nishizawa, T, Yamashita, K, Ishitani, R, Nureki, O. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the facilitative diffusion mechanism by SemiSWEET transporter
Nat Commun, 6, 2015
|
|
