6KBM
 
 | | Crystal structure of Vac8 bound to Atg13 | | Descriptor: | Autophagy-related protein 13, Vacuolar protein 8 | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-06-25 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways.
Autophagy, 16, 2020
|
|
1LCP
 
 | |
6LBX
 
 | | Crystal structure of HER2 Domain IV and Rb-H2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor tyrosine-protein kinase erbB-2, Repebody (Rb-H2) | | Authors: | Cho, H.S, Cha, J.S. | | Deposit date: | 2019-11-15 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Computationally-guided design and affinity improvement of a protein binder targeting a specific site on HER2
Comput Struct Biotechnol J, 19, 2021
|
|
7CM0
 
 | | Crystal structure of a glutaminyl cyclase in complex with NHV-1009 | | Descriptor: | 1-(cyclopentylmethyl)-1-[3-methoxy-4-(2-morpholin-4-ylethoxy)phenyl]-3-[3-(5-methylimidazol-1-yl)propyl]urea, Glutaminyl-peptide cyclotransferase, ZINC ION | | Authors: | Lee, J.W, Song, J.Y, Jang, T.H, Ha, J.H. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of highly potent human glutaminyl cyclase (QC) inhibitors as anti-Alzheimer's agents by the combination of pharmacophore-based and structure-based design.
Eur.J.Med.Chem., 226, 2021
|
|
4QBS
 
 | |
4QBR
 
 | |
4QBQ
 
 | | Crystal structure of DNMT3a ADD domain bound to H3 peptide | | Descriptor: | DNA (cytosine-5)-methyltransferase 3A, Histone H3, ZINC ION | | Authors: | Li, H, Patel, D.J. | | Deposit date: | 2014-05-08 | | Release date: | 2015-05-13 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Engineering of a histone-recognition domain in a de novo DNA methyltransferase alters the epigenetic landscape of ESCs
To be Published
|
|
3H0T
 
 | | Hepcidin-Fab complex | | Descriptor: | Fab fragment, Heavy chain, Light chain, ... | | Authors: | Syed, R, Li, V. | | Deposit date: | 2009-04-10 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Hepcidin revisited, disulfide connectivity, dynamics, and structure.
J.Biol.Chem., 284, 2009
|
|
5YI5
 
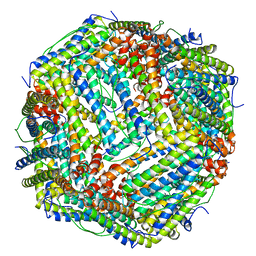 | | human ferritin mutant - E-helix deletion | | Descriptor: | Ferritin heavy chain | | Authors: | Lee, S.G, Yoon, H.R, Ahn, B.J, Jeong, H, Hyun, J, Jung, Y, Kim, H. | | Deposit date: | 2017-10-02 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Four-fold Channel-Nicked Human Ferritin Nanocages for Active Drug Loading and pH-Responsive Drug Release
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
2XHS
 
 | |
2Q8B
 
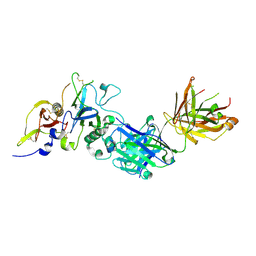 | | Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody | | Descriptor: | 1F9 heavy chain, 1F9 light chain, Apical membrane antigen 1 | | Authors: | Gupta, A, Murphy, V.J, Anders, R.F, Batchelor, A.H. | | Deposit date: | 2007-06-10 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Malaria Antigen AMA1 in Complex with a Growth-Inhibitory Antibody
Plos Pathog., 3, 2007
|
|
2IJN
 
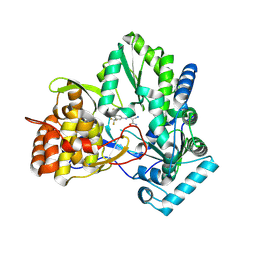 | | Isothiazoles as active-site inhibitors of HCV NS5B polymerase | | Descriptor: | (2R,3R)-3-{[3,5-BIS(TRIFLUOROMETHYL)PHENYL]AMINO}-2-CYANO-3-THIOXOPROPANAMIDE, RNA polymerase NS5B | | Authors: | Yan, S, Yao, N. | | Deposit date: | 2006-09-29 | | Release date: | 2006-11-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isothiazoles as active-site inhibitors of HCV NS5B polymerase
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2LM4
 
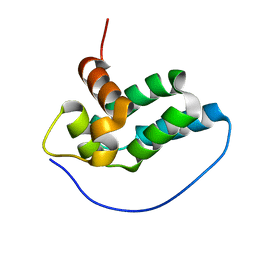 | | Solution NMR Structure of mitochondrial succinate dehydrogenase assembly factor 2 from Saccharomyces cerevisiae, Northeast Structural Genomics Consortium Target YT682A | | Descriptor: | Succinate dehydrogenase assembly factor 2, mitochondrial | | Authors: | Eletsky, A, Winge, D.R, Lee, H, Lee, D, Kohan, E, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2011-11-22 | | Release date: | 2012-01-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of yeast succinate dehydrogenase flavinylation factor sdh5 reveals a putative sdh1 binding site.
Biochemistry, 51, 2012
|
|
7KNW
 
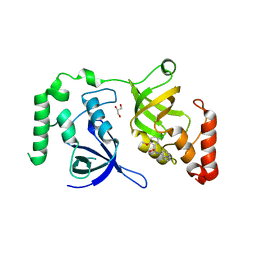 | | Crystal structure of SND1 in complex with C-26-A2 | | Descriptor: | 5-chloro-2-methoxy-N-([1,2,4]triazolo[1,5-a]pyridin-8-yl)benzene-1-sulfonamide, GLYCEROL, Staphylococcal nuclease domain-containing protein 1 | | Authors: | Kang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis.
Nat Cancer, 3, 2022
|
|
7KNX
 
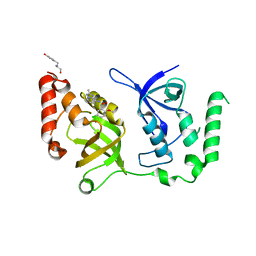 | | Crystal structure of SND1 in complex with C-26-A6 | | Descriptor: | 5-chloro-2-methoxy-N-(2-methyl[1,2,4]triazolo[1,5-a]pyridin-8-yl)benzene-1-sulfonamide, GLYCEROL, Staphylococcal nuclease domain-containing protein 1, ... | | Authors: | Kang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis.
Nat Cancer, 3, 2022
|
|
2LYQ
 
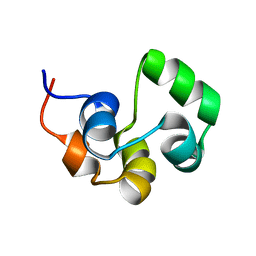 | | NOE-based 3D structure of the monomeric intermediate of CylR2 at 262K (-11 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYS
 
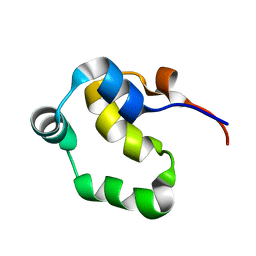 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 257K (-16 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYR
 
 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 259K (-14 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYP
 
 | | NOE-based 3D structure of the monomer of CylR2 in equilibrium with predissociated homodimer at 266K (-7 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYJ
 
 | | NOE-based 3D structure of the CylR2 homodimer at 298K | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Giller, K, Becker, S, Zweckstetter, M, Schwieters, C.D. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYK
 
 | | NOE-based 3D structure of the CylR2 homodimer at 270K (-3 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
4FJO
 
 | | Structure of the Rev1 CTD-Rev3/7-Pol kappa RIR complex | | Descriptor: | DNA polymerase kappa, DNA polymerase zeta catalytic subunit, DNA repair protein REV1, ... | | Authors: | Wojtaszek, J, Lee, C.-J, Zhou, P. | | Deposit date: | 2012-06-11 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.718 Å) | | Cite: | Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric Pol zeta and Pol kappa
J.Biol.Chem., 287, 2012
|
|
4EN1
 
 | |
2K21
 
 | | NMR structure of human KCNE1 in LMPG micelles at pH 6.0 and 40 degree C | | Descriptor: | Potassium voltage-gated channel subfamily E member | | Authors: | Kang, C, Tian, C, Sonnichsen, F.D, Smith, J.A, Meiler, J, George, A.L, Vanoye, C.G, Sanders, C.R, Kim, H. | | Deposit date: | 2008-03-19 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel.
Biochemistry, 47, 2008
|
|
2LYL
 
 | | NOE-based 3D structure of the predissociated homodimer of CylR2 in equilibrium with monomer at 266K (-7 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
