1F2C
 
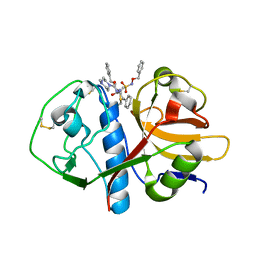 | | CRYSTAL STRUCTURE ANALYSIS OF CRYZAIN BOUND TO VINYL SULFONE DERIVED INHIBITOR (IV) | | Descriptor: | 3-[[N-[4-METHYL-PIPERAZINYL]CARBONYL]-PHENYLALANINYL-AMINO]-5-PHENYL-PENTANE-1-SULFONIC ACID BENZYLOXY-AMIDE, CRUZAIN | | Authors: | Brinen, L.S, Hansell, E, Roush, W.R, McKerrow, J.H, Fletterick, R.J. | | Deposit date: | 2000-05-23 | | Release date: | 2000-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A target within the target: probing cruzain's P1' site to define structural determinants for the Chagas' disease protease.
Structure Fold.Des., 8, 2000
|
|
1F29
 
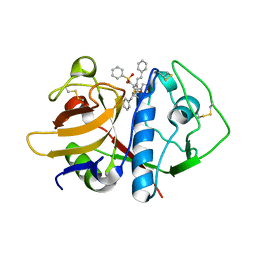 | | CRYSTAL STRUCTURE ANALYSIS OF CRUZAIN BOUND TO A VINYL SULFONE DERIVED INHIBITOR (I) | | Descriptor: | 3-[[N-[MORPHOLIN-N-YL]-CARBONYL]-PHENYLALANINYL-AMINO]-5- PHENYL-PENTANE-1-SULFONYLBENZENE, CRUZAIN | | Authors: | Brinen, L.S, Hansell, E, Roush, W.R, McKerrow, J.H, Fletterick, R.J. | | Deposit date: | 2000-05-23 | | Release date: | 2000-07-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A target within the target: probing cruzain's P1' site to define structural determinants for the Chagas' disease protease.
Structure Fold.Des., 8, 2000
|
|
2JSF
 
 | |
5WQ6
 
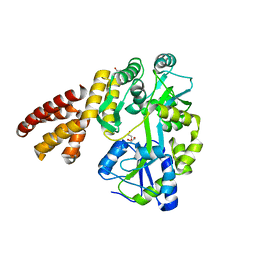 | | Crystal Structure of hMNDA-PYD with MBP tag | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MBP tagged hMNDA-PYD, ... | | Authors: | Jin, T.C, Xiao, T.S. | | Deposit date: | 2016-11-23 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Design of an expression system to enhance MBP-mediated crystallization
Sci Rep, 7, 2017
|
|
5WPZ
 
 | | Crystal structure of MNDA PYD with MBP tag | | Descriptor: | MBP-hMNDA-PYD, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T.C, Xiao, T.S. | | Deposit date: | 2016-11-22 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of an expression system to enhance MBP-mediated crystallization
Sci Rep, 7, 2017
|
|
8JSL
 
 | | The structure of EBOV L-VP35-RNA complex | | Descriptor: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, The leader sequence of EBOV, ... | | Authors: | Qi, P, Yi, S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Molecular mechanism of de novo replication by the Ebola virus polymerase.
Nature, 622, 2023
|
|
8JSM
 
 | | The structure of EBOV L-VP35-RNA complex (conformation 1) | | Descriptor: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, The leader sequence of EBOV genome., ... | | Authors: | Qi, P, Yi, S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular mechanism of de novo replication by the Ebola virus polymerase.
Nature, 622, 2023
|
|
8JSN
 
 | | The structure of EBOV L-VP35-RNA complex (conformation 2) | | Descriptor: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, The leader sequence of EBOV genome, ... | | Authors: | Qi, P, Yi, S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular mechanism of de novo replication by the Ebola virus polymerase.
Nature, 622, 2023
|
|
8JT8
 
 | | Crystal structure of 5-HT2AR in complex with (R)-IHCH-7179 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(4-fluorophenyl)-4-[(7R)-2,5,11-triazatetracyclo[7.6.1.0^2,7.0^12,16]hexadeca-1(15),9,12(16),13-tetraen-5-yl]butan-1-one, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Chen, Z, Fan, L, Wang, S. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Flexible scaffold-based cheminformatics approach for polypharmacological drug design.
Cell, 187, 2024
|
|
8JT6
 
 | | 5-HT1A-Gi in complex with compound (R)-IHCH-7179 | | Descriptor: | 1-(4-fluorophenyl)-4-[(7R)-2,5,11-triazatetracyclo[7.6.1.0^2,7.0^12,16]hexadeca-1(15),9,12(16),13-tetraen-5-yl]butan-1-one, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Chen, Z, Xu, P, Huang, S, Xu, H.E, Wang, S. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Flexible scaffold-based cheminformatics approach for polypharmacological drug design.
Cell, 187, 2024
|
|
5H7N
 
 | | Crystal structure of human NLRP12-PYD with a MBP tag | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NLRP12-PYD with MBP tag, ... | | Authors: | Jin, T.C, Xiao, T.S. | | Deposit date: | 2016-11-19 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Design of an expression system to enhance MBP-mediated crystallization
Sci Rep, 7, 2017
|
|
5H7Q
 
 | | Crystal structure of human MNDA PYD domain with MBP tag | | Descriptor: | ACETATE ION, MNDA PYD domain with MBP tag, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T.C, Xiao, T.S. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Design of an expression system to enhance MBP-mediated crystallization
Sci Rep, 7, 2017
|
|
6CN8
 
 | | High-resolution structure of ClpC1-NTD binding to Rufomycin-I | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC1, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Abad-Zapatero, C, Wolf, N.W. | | Deposit date: | 2018-03-07 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-Resolution Structure of ClpC1-Rufomycin and Ligand Binding Studies Provide a Framework to Design and Optimize Anti-Tuberculosis Leads.
Acs Infect Dis., 5, 2019
|
|
5ZOF
 
 | | Crystal Structure of D181A/R192F hFen1 in complex with DNA | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*G)-3'), DNA (5'-D(*CP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*G)-3'), DNA (5'-D(*GP*CP*CP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
5ZOG
 
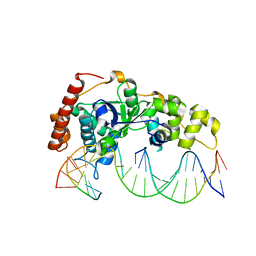 | | Crystal Structure of R192F hFen1 in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*G)-3'), ... | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
8ID6
 
 | | Cryo-EM structure of the oleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID4
 
 | | Cryo-EM structure of the linoleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID9
 
 | | Cryo-EM structure of the eicosapentaenoic acid bound GPR120-Gi complex | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID3
 
 | | Cryo-EM structure of the 9-hydroxystearic acid bound GPR120-Gi complex | | Descriptor: | 9-Hydroxyoctadecanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID8
 
 | | Cryo-EM structure of the TUG891 bound GPR120-Gi complex | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
6A50
 
 | | structure of benzoylformate decarboxylases in complex with cofactor TPP | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, benzoylformate decarboxylases | | Authors: | Guo, Y, Wang, S, Nie, Y, Li, S. | | Deposit date: | 2018-06-21 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Synthetic Pathway for Acetyl-Coenzyme A Biosynthesis
Nat Commun, 2019
|
|
1R6H
 
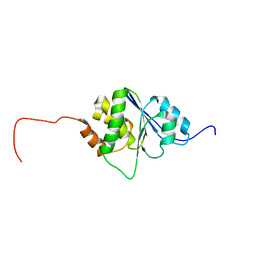 | | Solution Structure of human PRL-3 | | Descriptor: | protein tyrosine phosphatase type IVA, member 3 isoform 1 | | Authors: | Kozlov, G, Gehring, K, Ekiel, I. | | Deposit date: | 2003-10-15 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into Molecular Function of the Metastasis-associated Phosphatase PRL-3.
J.Biol.Chem., 279, 2004
|
|
8YKY
 
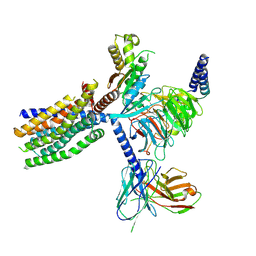 | | Structure of human class T GPCR TAS2R14-Ggustducin complex with agonist 28.1 | | Descriptor: | 4-methyl-N-[(2M)-2-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)pyrimidin-2-amine, CHOLESTEROL, G alpha gustducin protein, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-03-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQP
 
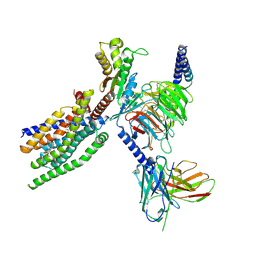 | | Structure of human class T GPCR TAS2R14-Gustducin complex with Aristolochic acid A. | | Descriptor: | 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQN
 
 | | Structure of human class T GPCR TAS2R14-DNGi complex with Aristolochic acid A. | | Descriptor: | 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
