6J3G
 
 | | Crystal structure of an apo form of the glutathione S-transferase, CsGST83044, of Ceriporiopsis subvermispora | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glutathione S-transferase, ... | | Authors: | Osman, W.H.W, Mikami, B, Saka, N, Kondo, K, Nagata, T, Katahira, M. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of key residues for activities of atypical glutathione S-transferase of Ceriporiopsis subvermispora, a selective degrader of lignin in woody biomass, by crystallography and functional mutagenesis.
Int.J.Biol.Macromol., 132, 2019
|
|
3FDW
 
 | | Crystal structure of a C2 domain from human synaptotagmin-like protein 4 | | Descriptor: | Synaptotagmin-like protein 4 | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-26 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a C2 domain from human synaptotagmin-like protein 4
To be Published
|
|
6IYZ
 
 | | Structural basis for activity of TRIC counter-ion channels in calcium release | | Descriptor: | CHLORIDE ION, Trimeric intracellular cation channel type A | | Authors: | Wang, X.H, Zeng, Y, Qin, L, Gao, F, Su, M, Chen, Y.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6J54
 
 | | Cryo-EM structure of the mammalian E-state ATP synthase FO section | | Descriptor: | ATP synthase membrane subunit 6.8PL, ATP synthase membrane subunit DAPIT, ATP synthase peripheral stalk-membrane subunit b, ... | | Authors: | Gu, J, Zhang, L, Yi, J, Yang, M. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Science, 364, 2019
|
|
8B3Q
 
 | |
3F1P
 
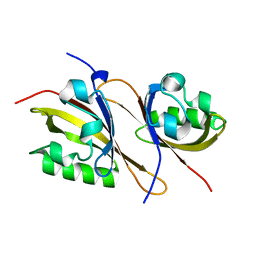 | | Crystal structure of a high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Scheuermann, T.H, Tomchick, D.R, Machius, M, Guo, Y, Bruick, R.K, Gardner, K.H. | | Deposit date: | 2008-10-28 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3FER
 
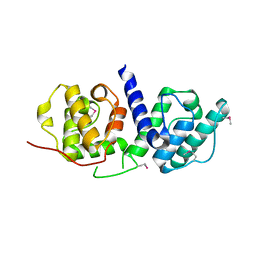 | | Crystal structure of n-terminal actin-binding domain from human filamin b (tandem ch-domains). northeast structural genomics consortium target hr5571a. | | Descriptor: | ACETIC ACID, Filamin-B | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, R, Shastry, R, Sahdev, S, Ciccosanti, C, Xiao, R, Everett, J.K, Huang, Y, Acton, T, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-11-30 | | Release date: | 2009-01-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of n-terminal actin-binding domain from human filamin b (tandem ch-domains). northeast structural genomics consortium target hr5571a.
To be Published
|
|
3F2H
 
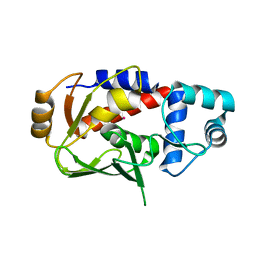 | | Crystal structure of the mercury-bound form of MerB mutant C160S, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase, MERCURY (II) ION | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-29 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
8B3P
 
 | |
3F3K
 
 | | The structure of uncharacterized protein YKR043C from Saccharomyces cerevisiae. | | Descriptor: | GLYCEROL, Uncharacterized protein YKR043C | | Authors: | Cuff, M, Xu, X, Cui, H, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and activity of the metal-independent fructose-1,6-bisphosphatase YK23 from Saccharomyces cerevisiae.
J.Biol.Chem., 285, 2010
|
|
6J07
 
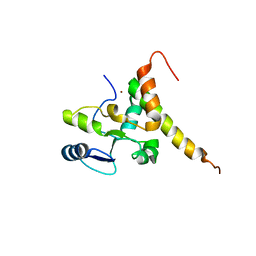 | | Crystal structure of human TERB2 and TERB1 | | Descriptor: | Telomere repeats-binding bouquet formation protein 1, Telomere repeats-binding bouquet formation protein 2, ZINC ION | | Authors: | Wang, Y, Chen, Y, Wu, J, Huang, C, Lei, M. | | Deposit date: | 2018-12-21 | | Release date: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (3.298 Å) | | Cite: | The meiotic TERB1-TERB2-MAJIN complex tethers telomeres to the nuclear envelope.
Nat Commun, 10, 2019
|
|
8B3O
 
 | |
6J0V
 
 | | Crystal Structure of Yeast Rtt107 | | Descriptor: | Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.314 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
8AOV
 
 | |
6J9Q
 
 | | Crystal structure of Trypanosoma brucei gambiense glycerol kinase complex with AMP-PNP. | | Descriptor: | GLYCEROL, Glycerol kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Balogun, E.O, Chishima, T, Ichinose, M, Inaoka, D.K, Kido, Y, Ibrahim, B, de Koning, H, McKerrow, J.H, Watanabe, Y, Nozaki, T, Michels, P.A.M, Harada, S, Kita, K, Shiba, T. | | Deposit date: | 2019-01-24 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Reaction mechanism of the reverse reaction of African human trypanosomes glycerol kinase.
To Be Published
|
|
6JB0
 
 | | Crystal structure of ABC transporter alpha-glycoside-binding mutant protein W287A in complex with trehalose | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ABC transporter, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-25 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
6J5A
 
 | | Cryo-EM structure of the mammalian DP-state ATP synthase FO section | | Descriptor: | ATP synthase membrane subunit 6.8PL, ATP synthase membrane subunit DAPIT, ATP synthase peripheral stalk-membrane subunit b, ... | | Authors: | Gu, J, Zhang, L, Yi, J, Yang, M. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Science, 364, 2019
|
|
3F7T
 
 | | Structure of active IspH shows a novel fold with a [3Fe-4S] cluster in the catalytic centre | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER, PHOSPHATE ION, ... | | Authors: | Graewert, T, Eppinger, J, Rohdich, F, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2008-11-10 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of active IspH enzyme from Escherichia coli provides mechanistic insights into substrate reduction.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3FJ9
 
 | |
3FKZ
 
 | | X-ray structure of the non covalent swapped form of the S16G/T17N/A19P/A20S/K31C/S32C mutant of bovine pancreatic ribonuclease | | Descriptor: | Ribonuclease pancreatic | | Authors: | Merlino, A, Russo Krauss, I, Perillo, M, Mattia, C.A, Ercole, C, Picone, D, Vergara, A, Sica, F. | | Deposit date: | 2008-12-18 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Toward an antitumor form of bovine pancreatic ribonuclease: The crystal structure of three noncovalent dimeric mutants
Biopolymers, 91, 2009
|
|
4UXG
 
 | | Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fibre protein gp34, R32 native crystal | | Descriptor: | LARGE TAIL FIBER PROTEIN P34 | | Authors: | Granell, M, Alvira, S, Garcia-Doval, C, Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34.
Viruses, 9, 2017
|
|
6J3H
 
 | | Crystal structure of the glutathione S-transferase, CsGST83044, of Ceriporiopsis subvermispora in complex with glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase | | Authors: | Osman, W.H.W, Mikami, B, Saka, N, Kondo, K, Nagata, T, Katahira, M. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identification of key residues for activities of atypical glutathione S-transferase of Ceriporiopsis subvermispora, a selective degrader of lignin in woody biomass, by crystallography and functional mutagenesis.
Int.J.Biol.Macromol., 132, 2019
|
|
8AVI
 
 | | Crystal structure of IsdG from Bacillus cereus in complex with heme | | Descriptor: | 1,3-PROPANDIOL, 1,4-BUTANEDIOL, Heme-degrading monooxygenase, ... | | Authors: | Andersen, H.K, Hammerstad, M, Hersleth, H.-P. | | Deposit date: | 2022-08-26 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Diversity of Homologous Oxidoreductases-Tuning of Substrate Specificity by a FAD-Stacking Residue for Iron Acquisition and Flavodoxin Reduction.
Antioxidants, 12, 2023
|
|
1DLF
 
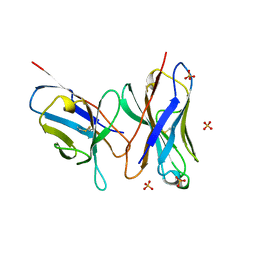 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE FV FRAGMENT FROM AN ANTI-DANSYL SWITCH VARIANT ANTIBODY IGG2A(S) CRYSTALLIZED AT PH 5.25 | | Descriptor: | ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S), SULFATE ION | | Authors: | Nakasako, M, Takahashi, H, Shimada, I, Arata, Y. | | Deposit date: | 1998-07-14 | | Release date: | 1999-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody.
J.Mol.Biol., 291, 1999
|
|
6J4W
 
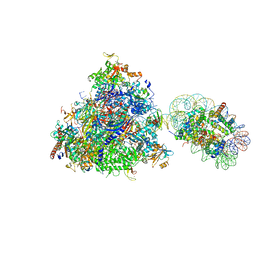 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
