4FM6
 
 | |
3NU6
 
 | |
3NUO
 
 | |
3NU9
 
 | |
3NUJ
 
 | |
4FL8
 
 | | HIV-1 protease complexed with gem-diol-amine tetrahedral intermediate | | 分子名称: | CHLORIDE ION, GLYCEROL, HIV-1 protease, ... | | 著者 | Tie, Y.F, Shen, C.H, Weber, I.T. | | 登録日 | 2012-06-14 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Capturing the Reaction Pathway in Near-Atomic-Resolution Crystal Structures of HIV-1 Protease.
Biochemistry, 51, 2012
|
|
4FLG
 
 | | HIV-1 protease mutant I47V complexed with reaction intermediate | | 分子名称: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | 著者 | Yu, X, Shen, C.H, Weber, I.T. | | 登録日 | 2012-06-14 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Capturing the Reaction Pathway in Near-Atomic-Resolution Crystal Structures of HIV-1 Protease.
Biochemistry, 51, 2012
|
|
4HDP
 
 | | Crystal Structure of HIV-1 protease mutants I50V complexed with inhibitor GRL-0519 | | 分子名称: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, HIV-1 Protease, ... | | 著者 | Shen, C.H, Zhang, H, Weber, I.T. | | 登録日 | 2012-10-02 | | 公開日 | 2013-08-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease.
J.Med.Chem., 56, 2013
|
|
4HE9
 
 | | Crystal Structure of HIV-1 protease mutants I54M complexed with inhibitor GRL-0519 | | 分子名称: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, HIV-1 protease, ... | | 著者 | Shen, C.H, Zhang, H, Weber, I.T. | | 登録日 | 2012-10-03 | | 公開日 | 2013-08-21 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.06 Å) | | 主引用文献 | Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease.
J.Med.Chem., 56, 2013
|
|
4J5J
 
 | |
3NU5
 
 | | Crystal Structure of HIV-1 Protease Mutant I50V with Antiviral Drug Amprenavir | | 分子名称: | ACETATE ION, CHLORIDE ION, SODIUM ION, ... | | 著者 | Wang, Y.-F, Shen, C.H, Weber, I.T. | | 登録日 | 2010-07-06 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
4HDB
 
 | | Crystal Structure of HIV-1 protease mutants D30N complexed with inhibitor GRL-0519 | | 分子名称: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, HIV-1 Protease, ... | | 著者 | Zhang, H, Wang, Y.-F, Shen, C.H, Agniswamy, J, Weber, I.T. | | 登録日 | 2012-10-02 | | 公開日 | 2013-08-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease.
J.Med.Chem., 56, 2013
|
|
4Z4X
 
 | |
4Z50
 
 | | Crystal Structure of Multidrug Resistant HIV-1 Protease Clinical Isolate PR20D25N with Tucked Flap | | 分子名称: | CHLORIDE ION, GLYCEROL, Protease, ... | | 著者 | Agniswamy, J, Shen, C.-H, Weber, I.T. | | 登録日 | 2015-04-02 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Conformational variation of an extreme drug resistant mutant of HIV protease.
J.Mol.Graph.Model., 62, 2015
|
|
6UBI
 
 | |
6UCE
 
 | |
6UCF
 
 | |
3NU3
 
 | | Wild Type HIV-1 Protease with Antiviral Drug Amprenavir | | 分子名称: | CHLORIDE ION, GLYCEROL, Protease, ... | | 著者 | Wang, Y.-F, Kovalevsky, A.Y, Weber, I.T. | | 登録日 | 2010-07-06 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.02 Å) | | 主引用文献 | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
3NU4
 
 | | Crystal Structure of HIV-1 Protease Mutant V32I with Antiviral Drug Amprenavir | | 分子名称: | CHLORIDE ION, SODIUM ION, protease, ... | | 著者 | Wang, Y.-F, Kovalevsky, A.Y, Weber, I.T. | | 登録日 | 2010-07-06 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
6E5P
 
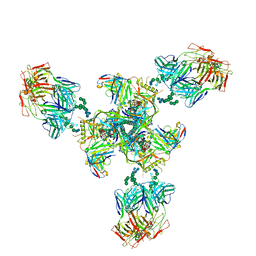 | | Backbone model based on cryo-EM map at 8.5 A of domain-swapped, glycan-reactive, neutralizing antibody 2G12 bound to HIV-1 Env BG505 DS-SOSIP, which was also bound to CD4-binding site antibody VRC03 | | 分子名称: | 2G12 Light chain, 2G12 heavy chain, Envelope glycoprotein gp120, ... | | 著者 | Acharya, P, Kwong, P.D. | | 登録日 | 2018-07-21 | | 公開日 | 2019-02-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (8.8 Å) | | 主引用文献 | Structural Survey of Broadly Neutralizing Antibodies Targeting the HIV-1 Env Trimer Delineates Epitope Categories and Characteristics of Recognition.
Structure, 27, 2019
|
|
7SG5
 
 | |
7SG6
 
 | |
6BF4
 
 | |
6OT1
 
 | |
6OSY
 
 | |
