1YKR
 
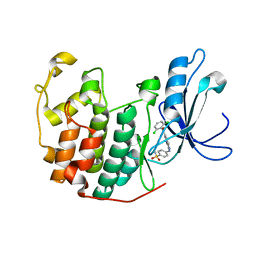 | | Crystal structure of cdk2 with an aminoimidazo pyridine inhibitor | | 分子名称: | 4-{[6-(2,6-DICHLOROBENZOYL)IMIDAZO[1,2-A]PYRIDIN-2-YL]AMINO}BENZENESULFONAMIDE, Cell division protein kinase 2 | | 著者 | Hamdouchi, C, Zhong, B, Mendoza, J, Jaramillo, C, Zhang, F, Brooks, H.B. | | 登録日 | 2005-01-18 | | 公開日 | 2006-01-24 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-based design of a new class of highly selective aminoimidazo[1,2-a]pyridine-based inhibitors of cyclin dependent kinases
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1PYE
 
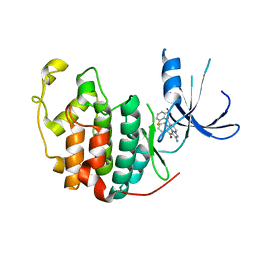 | | Crystal structure of CDK2 with inhibitor | | 分子名称: | Cell division protein kinase 2, [2-AMINO-6-(2,6-DIFLUORO-BENZOYL)-IMIDAZO[1,2-A]PYRIDIN-3-YL]-PHENYL-METHANONE | | 著者 | Zhang, F, Hamdouchi, C. | | 登録日 | 2003-07-08 | | 公開日 | 2004-07-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The discovery of a new structural class of cyclin-dependent kinase inhibitors, aminoimidazo[1,2-a]pyridines.
MOL.CANCER THER., 3, 2004
|
|
8FW5
 
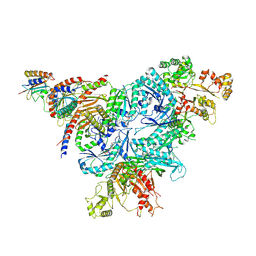 | | Chimeric HsGATOR1-SpGtr-SpLam complex | | 分子名称: | ALUMINUM FLUORIDE, GATOR complex protein DEPDC5, GATOR complex protein NPRL2, ... | | 著者 | Tettoni, S.D, Egri, S.B, Doxsey, D.D, Ouch, C, Chang, J, Song, K, Xu, C, Shen, K. | | 登録日 | 2023-01-20 | | 公開日 | 2023-07-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Structure of the Schizosaccharomyces pombe Gtr-Lam complex reveals evolutionary divergence of mTORC1-dependent amino acid sensing.
Structure, 31, 2023
|
|
8D8O
 
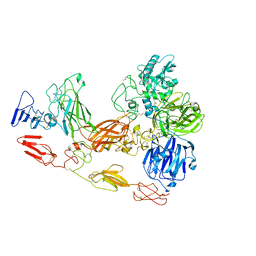 | | Cryo-EM structure of substrate unbound PAPP-A | | 分子名称: | Pappalysin-1, ZINC ION | | 著者 | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | 登録日 | 2022-06-08 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
7UFG
 
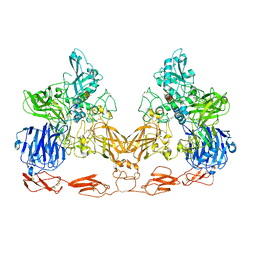 | | Cryo-EM structure of PAPP-A in complex with IGFBP5 | | 分子名称: | Insulin-like growth factor-binding protein 5, Pappalysin-1, ZINC ION | | 著者 | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | 登録日 | 2022-03-22 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
7N85
 
 | | Inner ring spoke from the isolated yeast NPC | | 分子名称: | Nucleoporin ASM4, Nucleoporin NIC96, Nucleoporin NSP1, ... | | 著者 | Akey, C.W, Rout, M.P, Ouch, C, Echevarria, I, Fernandez-Martinez, J, Nudelman, I. | | 登録日 | 2021-06-13 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (7.6 Å) | | 主引用文献 | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
7N84
 
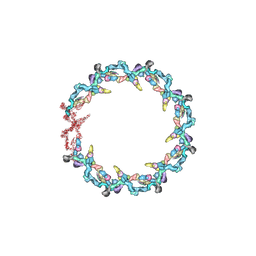 | | Double nuclear outer ring from the isolated yeast NPC | | 分子名称: | Nucleoporin 145c, Nucleoporin NUP120, Nucleoporin NUP133, ... | | 著者 | Akey, C.W, Rout, M.P, Ouch, C, Echevarria, I, Fernandez-Martinez, J, Nudelman, I. | | 登録日 | 2021-06-13 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (11.6 Å) | | 主引用文献 | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
8T9L
 
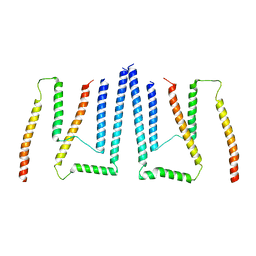 | | Pom34-Pom152 membrane attachment site yeast NPC | | 分子名称: | Nucleoporin POM152, Nucleoporin POM34 | | 著者 | Akey, C.W, Echeverria, I, Ouch, C, Fernandez-Martinez, J, Rout, M.P. | | 登録日 | 2023-06-24 | | 公開日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Implications of a multiscale structure of the yeast nuclear pore complex.
Mol.Cell, 83, 2023
|
|
8TJ5
 
 | | Inner spoke ring of the yeast NPC | | 分子名称: | Nucleoporin 59, Nucleoporin NIC96, Nucleoporin NSP1, ... | | 著者 | Akey, C.W, Echeverria, I, Ouch, C, Fernandez-Martinez, J, Rout, M.P. | | 登録日 | 2023-07-20 | | 公開日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (6.6 Å) | | 主引用文献 | Implications of a multiscale structure of the yeast nuclear pore complex.
Mol.Cell, 83, 2023
|
|
8TIE
 
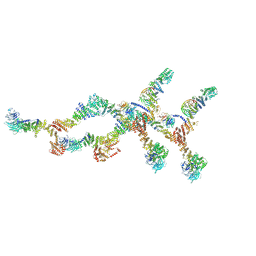 | | Double nuclear outer ring of Nup84-complexes from the yeast NPC | | 分子名称: | NUP133 isoform 1, NUP145 isoform 1, Nucleoporin NUP120, ... | | 著者 | Akey, C.W, Echeverria, I, Ouch, C, Fernandez-Martinez, J, Rout, M.P. | | 登録日 | 2023-07-19 | | 公開日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (8.1 Å) | | 主引用文献 | Implications of a multiscale structure of the yeast nuclear pore complex.
Mol.Cell, 83, 2023
|
|
5KW2
 
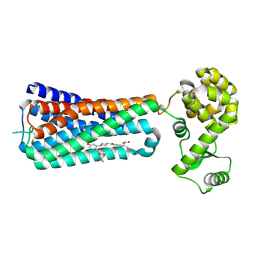 | | The extra-helical binding site of GPR40 and the structural basis for allosteric agonism and incretin stimulation | | 分子名称: | (3~{S})-3-cyclopropyl-3-[2-[1-[2-[2,2-dimethylpropyl-(6-methylpyridin-2-yl)carbamoyl]-5-methoxy-phenyl]piperidin-4-yl]-1-benzofuran-6-yl]propanoic acid, Free fatty acid receptor 1,Lysozyme,Free fatty acid receptor 1 | | 著者 | Ho, J.D, Chau, B, Rodgers, L, Lu, F, Wilbur, K.L, Otto, K.A, Chen, Y, Song, M, Riley, J.P, Yang, H.-C, Reynolds, N.A, Kahl, S.D, Lewis, A.P, Groshong, C, Madsen, R.E, Conners, K, Linswala, J.P, Gheyi, T, Saflor, M.D, Lee, M.R, Benach, J, Baker, K.A, Montrose-Rafizadeh, C, Genin, M.J, Miller, A.R, Hamdouchi, C. | | 登録日 | 2016-07-15 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Structural basis for GPR40 allosteric agonism and incretin stimulation.
Nat Commun, 9, 2018
|
|
3ZLS
 
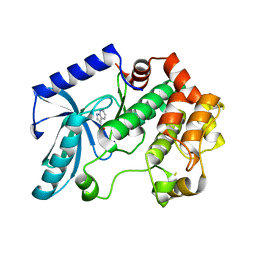 | | Crystal structure of MEK1 in complex with fragment 6 | | 分子名称: | 1H-PYRROLO[2,3-B]PYRIDINE-3-CARBOXYLIC ACID, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, SODIUM ION | | 著者 | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | 登録日 | 2013-02-04 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZM4
 
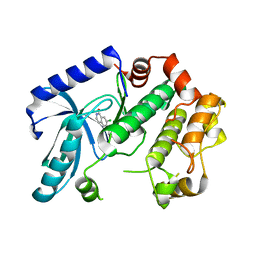 | | Crystal structure of MEK1 in complex with fragment 1 | | 分子名称: | 7-chloranyl-6-[(3S)-pyrrolidin-3-yl]oxy-2H-isoquinolin-1-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 | | 著者 | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | 登録日 | 2013-02-05 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZLW
 
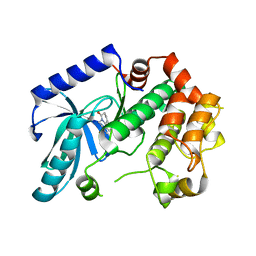 | | Crystal structure of MEK1 in complex with fragment 3 | | 分子名称: | (1R)-1-hydroxy-1-methyl-2,3,6,7-tetrahydro-1H,5H-pyrido[3,2,1-ij]quinolin-5-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 MAPK/ERK KINASE 1, MEK 1, ... | | 著者 | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | 登録日 | 2013-02-04 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZLY
 
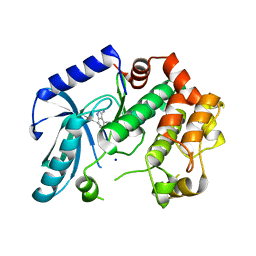 | | Crystal structure of MEK1 in complex with fragment 8 | | 分子名称: | 3-AMINO-1H-INDAZOLE-4-CARBONITRILE, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, SODIUM ION | | 著者 | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | 登録日 | 2013-02-04 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZLX
 
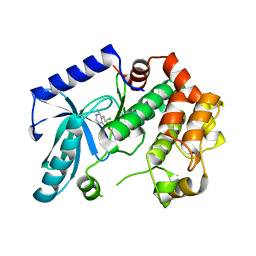 | | Crystal structure of MEK1 in complex with fragment 18 | | 分子名称: | 7-choro-6-[(3R)-pyrrolidin-3-ylmethoxy]isoquinolin-1(2H)-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 | | 著者 | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | 登録日 | 2013-02-04 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4XX5
 
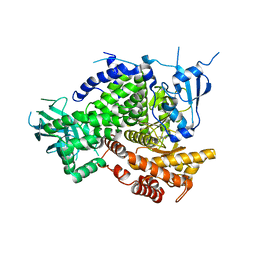 | | Structure of PI3K gamma in complex with an inhibitor | | 分子名称: | N-[5-(5-methoxypyridin-3-yl)-4,5,6,7-tetrahydro[1,3]thiazolo[5,4-c]pyridin-2-yl]acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | 著者 | Collier, P.N, Messersmith, D, Le Tiran, A, Bandarage, U.K, Boucher, C, Come, J, Cottrell, K.M, Damagnez, V, Doran, J.D, Griffith, J.P, Khare-Pandit, S, Krueger, E.B, Ledeboer, M.W, Ledford, B, Liao, Y, Mahajan, S, Moody, C.S, Wang, T, Xu, J, Aronov, A.M. | | 登録日 | 2015-01-29 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Structure of PI3K gamma in complex with an inhibitor
To Be Published
|
|
4XZ4
 
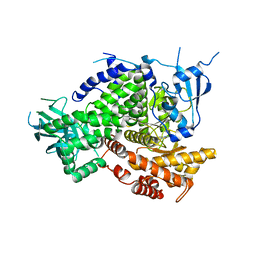 | | Structure of PI3K gamma in complex with an inhibitor | | 分子名称: | N-[5-(6-methoxypyrazin-2-yl)-4,5,6,7-tetrahydro[1,3]thiazolo[5,4-c]pyridin-2-yl]acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | 著者 | Collier, P.N, Messersmith, D, Le Tiran, A, Bandarage, U.K, Boucher, C, Come, J, Cottrell, K.M, Damagnez, V, Doran, J.D, Griffith, J.P, Khare-Pandit, S, Krueger, E.B, Ledeboer, M.W, Ledford, B, Liao, Y, Mahajan, S, Moody, C.S, Wang, T, Xu, J, Aronov, A.M. | | 登録日 | 2015-02-03 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of PI3K gamma in complex with an inhibitor
To Be Published
|
|
7T3A
 
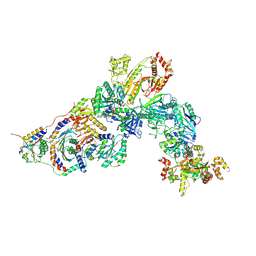 | | GATOR1-RAG-RAGULATOR - Inhibitory Complex | | 分子名称: | GATOR complex protein DEPDC5, GATOR complex protein NPRL2, GATOR complex protein NPRL3, ... | | 著者 | Egri, S.B, Shen, K. | | 登録日 | 2021-12-07 | | 公開日 | 2022-04-06 | | 最終更新日 | 2022-06-01 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Cryo-EM structures of the human GATOR1-Rag-Ragulator complex reveal a spatial-constraint regulated GAP mechanism.
Mol.Cell, 82, 2022
|
|
7T3B
 
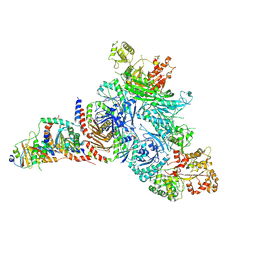 | | GATOR1-RAG-RAGULATOR - GAP Complex | | 分子名称: | ALUMINUM FLUORIDE, GATOR complex protein DEPDC5, GATOR complex protein NPRL2, ... | | 著者 | Egri, S.B, Shen, K. | | 登録日 | 2021-12-07 | | 公開日 | 2022-04-06 | | 最終更新日 | 2022-06-01 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structures of the human GATOR1-Rag-Ragulator complex reveal a spatial-constraint regulated GAP mechanism.
Mol.Cell, 82, 2022
|
|
7T3C
 
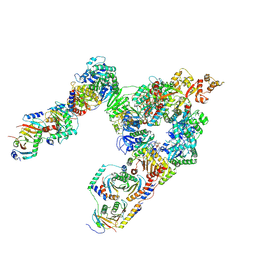 | | GATOR1-RAG-RAGULATOR - Dual Complex | | 分子名称: | ALUMINUM FLUORIDE, GATOR complex protein DEPDC5, GATOR complex protein NPRL2, ... | | 著者 | Egri, S.B, Shen, K. | | 登録日 | 2021-12-07 | | 公開日 | 2022-04-06 | | 最終更新日 | 2022-06-01 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Cryo-EM structures of the human GATOR1-Rag-Ragulator complex reveal a spatial-constraint regulated GAP mechanism.
Mol.Cell, 82, 2022
|
|
7N9F
 
 | | Structure of the in situ yeast NPC | | 分子名称: | Dynein light chain 1, cytoplasmic, Nucleoporin 145c, ... | | 著者 | Villa, E, Singh, D, Ludtke, S.J, Akey, C.W, Rout, M.P, Echeverria, I, Suslov, S. | | 登録日 | 2021-06-17 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (37 Å) | | 主引用文献 | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
4JMU
 
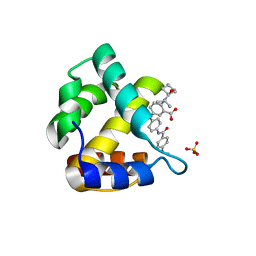 | | Crystal structure of HIV matrix residues 1-111 in complex with inhibitor | | 分子名称: | 5-{4-[(4-methoxybenzoyl)amino]phenoxy}-2-{[(trans-4-methylcyclohexyl)carbonyl](propan-2-yl)amino}benzoic acid, Gag-Pol polyprotein, SULFATE ION | | 著者 | Lemke, C.T. | | 登録日 | 2013-03-14 | | 公開日 | 2013-10-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Enantiomeric Atropisomers Inhibit HCV Polymerase and/or HIV Matrix: Characterizing Hindered Bond Rotations and Target Selectivity.
J.Med.Chem., 57, 2014
|
|
4JVQ
 
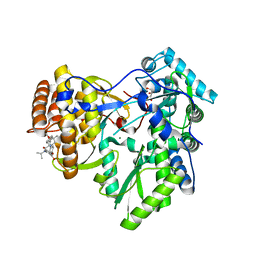 | | Crystal structure of hcv ns5b polymerase in complex with compound 9 | | 分子名称: | 5-{4-[(4-methoxybenzoyl)amino]phenoxy}-2-{[(trans-4-methylcyclohexyl)carbonyl](propan-2-yl)amino}benzoic acid, GLYCEROL, Genome polyprotein, ... | | 著者 | Coulombe, R. | | 登録日 | 2013-03-26 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Enantiomeric Atropisomers Inhibit HCV Polymerase and/or HIV Matrix: Characterizing Hindered Bond Rotations and Target Selectivity.
J.Med.Chem., 57, 2014
|
|
3BYM
 
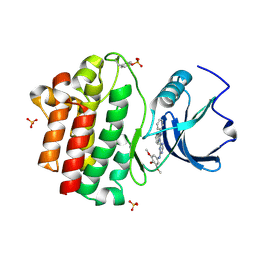 | | X-ray co-crystal structure aminobenzimidazole triazine 1 bound to Lck | | 分子名称: | N-phenyl-1-{4-[(3,4,5-trimethoxyphenyl)amino]-1,3,5-triazin-2-yl}-1H-benzimidazol-2-amine, Proto-oncogene tyrosine-protein kinase LCK, SULFATE ION | | 著者 | Huang, X. | | 登録日 | 2008-01-16 | | 公開日 | 2008-09-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-based design of novel 2-amino-6-phenyl-pyrimido[5',4':5,6]pyrimido[1,2-a]benzimidazol-5(6H)-ones as potent and orally active inhibitors of lymphocyte specific kinase (Lck): synthesis, SAR, and in vivo anti-inflammatory activity.
J.Med.Chem., 51, 2008
|
|
