3WJ2
 
 | | Crystal structure of ESTFA (FE-lacking apo form) | | 分子名称: | Carboxylesterase | | 著者 | Ohara, K, Unno, H, Oshima, Y, Furukawa, K, Fujino, N, Hirooka, K, Hemmi, H, Takahashi, S, Nishino, T, Kusunoki, M, Nakayama, T. | | 登録日 | 2013-10-03 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Structural insights into the low pH adaptation of a unique carboxylesterase from Ferroplasma: altering the pH optima of two carboxylesterases.
J.Biol.Chem., 289, 2014
|
|
3WJ1
 
 | | Crystal structure of SSHESTI | | 分子名称: | Carboxylesterase, octyl beta-D-glucopyranoside | | 著者 | Ohara, K, Unno, H, Oshima, Y, Furukawa, K, Fujino, N, Hirooka, K, Hemmi, H, Takahashi, S, Nishino, T, Kusunoki, M, Nakayama, T. | | 登録日 | 2013-10-03 | | 公開日 | 2014-07-30 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural insights into the low pH adaptation of a unique carboxylesterase from Ferroplasma: altering the pH optima of two carboxylesterases.
J.Biol.Chem., 289, 2014
|
|
5ZUI
 
 | | Crystal Structure of HSP104 from Chaetomium thermophilum | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Heat Shock Protein 104, SULFATE ION | | 著者 | Hanazono, Y, Inoue, Y, Noguchi, K, Yohda, M, Shinohara, K, Takeda, K, Miki, K. | | 登録日 | 2018-05-07 | | 公開日 | 2019-06-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | Split conformation of Chaetomium thermophilum Hsp104 disaggregase.
Structure, 2021
|
|
7CG3
 
 | | Staggered ring conformation of CtHsp104 (Hsp104 from Chaetomium Thermophilum) | | 分子名称: | Heat shock protein 104 | | 著者 | Inoue, Y, Hanazono, Y, Noi, K, Kawamoto, A, Kimatsuka, M, Harada, R, Takeda, K, Iwamasa, N, Shibata, K, Noguchi, K, Shigeta, Y, Namba, K, Ogura, T, Miki, K, Shinohara, K, Yohda, M. | | 登録日 | 2020-06-30 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Split conformation of Chaetomium thermophilum Hsp104 disaggregase.
Structure, 29, 2021
|
|
4P9N
 
 | | Crystal structure of sshesti PE mutant | | 分子名称: | Carboxylesterase | | 著者 | Unno, H. | | 登録日 | 2014-04-04 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Insights into the Low pH Adaptation of a Unique Carboxylesterase from Ferroplasma: ALTERING THE pH OPTIMA OF TWO CARBOXYLESTERASES.
J.Biol.Chem., 289, 2014
|
|
8YFY
 
 | | CRYSTAL STRUCTURE OF THE EST1 H274D MUTANT AT PH 4.2 | | 分子名称: | Carboxylesterase, octyl beta-D-glucopyranoside | | 著者 | Unno, H, Oshima, Y, Nishino, T, Nakayama, T, Kusunoki, M. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-08-28 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Lowering pH optimum of activity of SshEstI, a slightly alkaliphilic archaeal esterase of the hormone-sensitive lipase family.
J.Biosci.Bioeng., 138, 2024
|
|
8YFZ
 
 | | CRYSTAL STRUCTURE OF THE EST1 H274E MUTANT AT PH 4.2 | | 分子名称: | Carboxylesterase, octyl beta-D-glucopyranoside | | 著者 | Unno, H, Oshima, Y, Nishino, T, Nakayama, T, Kusunoki, M. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Lowering pH optimum of activity of SshEstI, a slightly alkaliphilic archaeal esterase of the hormone-sensitive lipase family.
J.Biosci.Bioeng., 2024
|
|
7YUZ
 
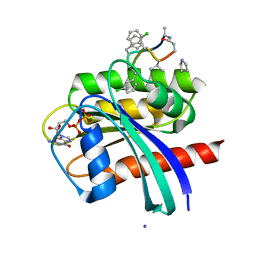 | | Human K-Ras G12D (GDP-bound) in complex with cyclic peptide inhibitor AP8784 | | 分子名称: | AP8784, GUANOSINE-5'-DIPHOSPHATE, IODIDE ION, ... | | 著者 | Irie, M, Fukami, T.A, Tanada, M, Ohta, A, Torizawa, T. | | 登録日 | 2022-08-18 | | 公開日 | 2023-07-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.878 Å) | | 主引用文献 | Validation of a New Methodology to Create Oral Drugs beyond the Rule of 5 for Intracellular Tough Targets.
J.Am.Chem.Soc., 145, 2023
|
|
7YV1
 
 | | Human K-Ras G12D (GDP-bound) in complex with cyclic peptide inhibitor LUNA18 and KA30L Fab | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, KA30L Fab H-chain, ... | | 著者 | Irie, M, Fukami, T.A, Matsuo, A, Saka, K, Nishimura, M, Saito, H, Torizawa, T, Tanada, M, Ohta, A. | | 登録日 | 2022-08-18 | | 公開日 | 2023-07-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.454 Å) | | 主引用文献 | Validation of a New Methodology to Create Oral Drugs beyond the Rule of 5 for Intracellular Tough Targets.
J.Am.Chem.Soc., 145, 2023
|
|
7DC8
 
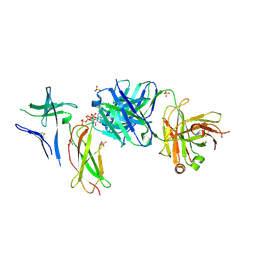 | | Crystal structure of Switch Ab Fab and hIL6R in complex with ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Interleukin-6 receptor subunit alpha, SULFATE ION, ... | | 著者 | Kadono, S, Fukami, T.A, Kawauchi, H, Torizawa, T, Mimoto, F. | | 登録日 | 2020-10-23 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.757 Å) | | 主引用文献 | Exploitation of Elevated Extracellular ATP to Specifically Direct Antibody to Tumor Microenvironment.
Cell Rep, 33, 2020
|
|
7DC7
 
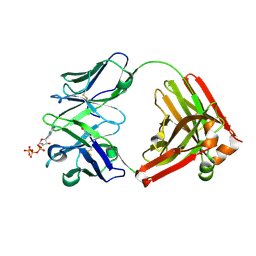 | | Crystal structure of D12 Fab-ATP complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, D12 Fab heavy chain, D12 Fab light chain | | 著者 | Kawauchi, H, Fukami, T.A, Tatsumi, K, Torizawa, T, Mimoto, F. | | 登録日 | 2020-10-23 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Exploitation of Elevated Extracellular ATP to Specifically Direct Antibody to Tumor Microenvironment.
Cell Rep, 33, 2020
|
|
8JJS
 
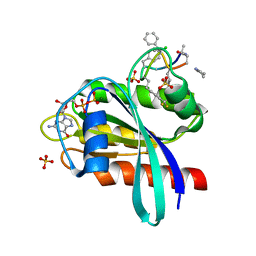 | | Human K-Ras G12D (GDP-bound) in complex with cyclic peptide inhibitor AP10343 | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, MAA-ILE-SAR-SAR-7T2-SAR-IAE-LEU-MEA-MLE-7TK, ... | | 著者 | Irie, M, Fukami, T.A, Tanada, M, Ohta, A, Torizawa, T. | | 登録日 | 2023-05-31 | | 公開日 | 2023-07-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.534 Å) | | 主引用文献 | Development of Orally Bioavailable Peptides Targeting an Intracellular Protein: From a Hit to a Clinical KRAS Inhibitor.
J.Am.Chem.Soc., 145, 2023
|
|
3WIG
 
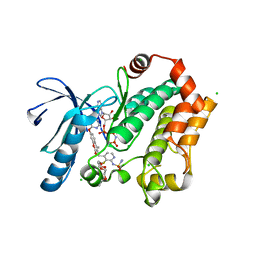 | | Human MEK1 kinase in complex with CH5126766 and MgAMP-PNP | | 分子名称: | CHLORIDE ION, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | 著者 | Lukacs, C.M, Janson, C, Schuck, V. | | 登録日 | 2013-09-12 | | 公開日 | 2014-06-04 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Disruption of CRAF-Mediated MEK Activation Is Required for Effective MEK Inhibition in KRAS Mutant Tumors
Cancer Cell, 25, 2014
|
|
3W0T
 
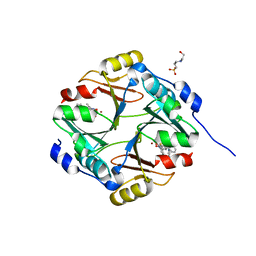 | |
3VW9
 
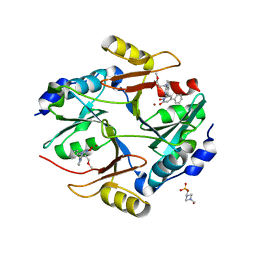 | | Human Glyoxalase I with an N-hydroxypyridone inhibitor | | 分子名称: | 1-hydroxy-6-[1-(3-methoxypropyl)-1H-pyrrolo[2,3-b]pyridin-5-yl]-4-phenylpyridin-2(1H)-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lactoylglutathione lyase, ... | | 著者 | Fukami, T.A, Irie, M, Matsuura, T. | | 登録日 | 2012-08-10 | | 公開日 | 2012-12-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Design and evaluation of azaindole-substituted N-hydroxypyridones as glyoxalase I inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3W0U
 
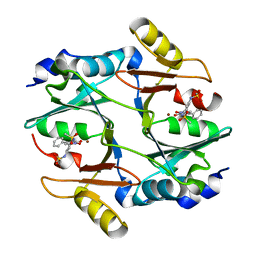 | | human Glyoxalase I with an N-hydroxypyridone inhibitor | | 分子名称: | Lactoylglutathione lyase, N-[3-(1-Hydroxy-6-oxo-4-phenyl-1,6-dihydro-pyridin-2-yl)-5-methanesulfonylamino-phenyl]-methanesulfonamide, ZINC ION | | 著者 | Fukami, T.A, Irie, M, Matsuura, T. | | 登録日 | 2012-11-02 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | N-Hydroxypyridone-based glyoxalase I inhibitors mimicking binding interactions of the substrate
to be published
|
|
8DOY
 
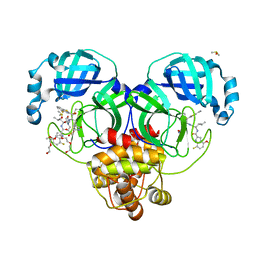 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-198 | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3C-like proteinase nsp5, ... | | 著者 | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | 登録日 | 2022-07-14 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Potent and biostable inhibitors of the main protease of SARS-CoV-2.
Iscience, 25, 2022
|
|
9FQI
 
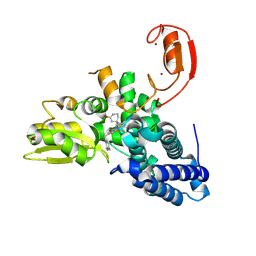 | | E3 ligase Cbl-b in complex with a lactam scaffold inhibitor (compound 7) | | 分子名称: | 8-[3-[(4~{R})-4-methyl-2-oxidanylidene-piperidin-4-yl]phenyl]-3-[[(3~{S})-3-methylpiperidin-1-yl]methyl]-5-(trifluoromethyl)-1$l^{4},7,8-triazabicyclo[4.3.0]nona-1(6),2,4-trien-9-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | 著者 | Schimpl, M. | | 登録日 | 2024-06-17 | | 公開日 | 2024-07-31 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.954 Å) | | 主引用文献 | Accelerated Discovery of Carbamate Cbl-b Inhibitors Using Generative AI Models and Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
9FQJ
 
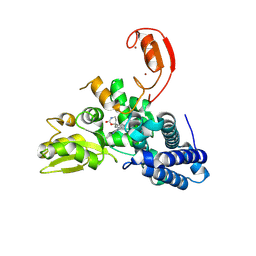 | | E3 ligase Cbl-b in complex with a carbamate scaffold inhibitor (compound 12) | | 分子名称: | 2-cyclopropyl-6-methyl-~{N}-[3-[(6~{S})-6-methyl-2-oxidanylidene-1,3-oxazinan-6-yl]phenyl]pyrimidine-4-carboxamide, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | 著者 | Schimpl, M. | | 登録日 | 2024-06-17 | | 公開日 | 2024-07-31 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.563 Å) | | 主引用文献 | Accelerated Discovery of Carbamate Cbl-b Inhibitors Using Generative AI Models and Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
9FQH
 
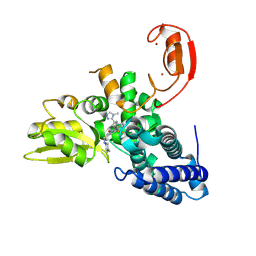 | | E3 ligase Cbl-b in complex with a triazolone core inhibitor (compound 1) | | 分子名称: | 8-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-3-[[(3~{S})-3-methylpiperidin-1-yl]methyl]-5-(trifluoromethyl)-1$l^{4},7,8-triazabicyclo[4.3.0]nona-1(6),2,4-trien-9-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | 著者 | Schimpl, M. | | 登録日 | 2024-06-17 | | 公開日 | 2024-07-31 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.786 Å) | | 主引用文献 | Accelerated Discovery of Carbamate Cbl-b Inhibitors Using Generative AI Models and Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
