8FY2
 
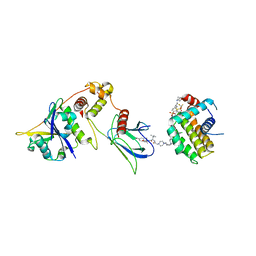 | | E3:PROTAC:target ternary complex structure (VCB/WH244/BCL-2) | | 分子名称: | Apoptosis regulator Bcl-2, Elongin-B, Elongin-C, ... | | 著者 | Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Ruben, E, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D, Olsen, S.K. | | 登録日 | 2023-01-25 | | 公開日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
8FY1
 
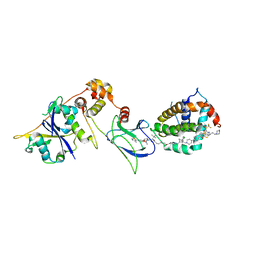 | | E3:PROTAC:target ternary complex structure (VCB/753b/BCL-2) | | 分子名称: | Apoptosis regulator Bcl-2, Elongin-B, Elongin-C, ... | | 著者 | Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D, Olsen, S.K. | | 登録日 | 2023-01-25 | | 公開日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
5H7R
 
 | |
5H7S
 
 | |
8FY0
 
 | | E3:PROTAC:target ternary complex structure (VCB/753b/BCL-xL) | | 分子名称: | Bcl-2-like protein 1, CACODYLIC ACID, Elongin-B, ... | | 著者 | Olsen, S.K, Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D. | | 登録日 | 2023-01-25 | | 公開日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
5DIN
 
 | |
6JX6
 
 | | Tetrameric form of Smac | | 分子名称: | Diablo homolog, mitochondrial | | 著者 | Sivaraman, J, Singh, S, Ng, J, Nayak, D. | | 登録日 | 2019-04-22 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.805 Å) | | 主引用文献 | Structural insights into a HECT-type E3 ligase AREL1 and its ubiquitination activitiesin vitro.
J.Biol.Chem., 294, 2019
|
|
6JX5
 
 | | Hect domain of AREL1 | | 分子名称: | Apoptosis-resistant E3 ubiquitin protein ligase 1 | | 著者 | Sivaraman, J, Singh, S, Ng, J, Nayak, D. | | 登録日 | 2019-04-22 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.402 Å) | | 主引用文献 | Structural insights into a HECT-type E3 ligase AREL1 and its ubiquitination activitiesin vitro.
J.Biol.Chem., 294, 2019
|
|
6WC6
 
 | |
8SEB
 
 | | Cryo-EM structure of a single loaded human UBA7-UBE2L6-ISG15 adenylate complex | | 分子名称: | ADENOSINE MONOPHOSPHATE, Ubiquitin-like modifier-activating enzyme 7, Ubiquitin-like protein ISG15, ... | | 著者 | Afsar, M, Jia, L, Ruben, E.A, Olsen, S.K. | | 登録日 | 2023-04-08 | | 公開日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | Cryo-EM structures of Uba7 reveal the molecular basis for ISG15 activation and E1-E2 thioester transfer.
Nat Commun, 14, 2023
|
|
8SE9
 
 | | Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex (Form 2) | | 分子名称: | ADENOSINE MONOPHOSPHATE, Ubiquitin-like modifier-activating enzyme 7, Ubiquitin-like protein ISG15, ... | | 著者 | Afsar, M, Jia, L, Ruben, E.A, Olsen, S.K. | | 登録日 | 2023-04-08 | | 公開日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures of Uba7 reveal the molecular basis for ISG15 activation and E1-E2 thioester transfer.
Nat Commun, 14, 2023
|
|
8SEA
 
 | | Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex (Form 1) | | 分子名称: | ADENOSINE MONOPHOSPHATE, Ubiquitin-like modifier-activating enzyme 7, Ubiquitin-like protein ISG15, ... | | 著者 | Afsar, M, Jia, L, Ruben, E.A, Olsen, S.K. | | 登録日 | 2023-04-08 | | 公開日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structures of Uba7 reveal the molecular basis for ISG15 activation and E1-E2 thioester transfer.
Nat Commun, 14, 2023
|
|
8SV8
 
 | | Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex from a composite map | | 分子名称: | ADENOSINE MONOPHOSPHATE, Ubiquitin-like modifier-activating enzyme 7, Ubiquitin-like protein ISG15, ... | | 著者 | Afsar, M, Jia, L, Ruben, E.A, Olsen, S.K. | | 登録日 | 2023-05-15 | | 公開日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.38 Å) | | 主引用文献 | Cryo-EM structures of Uba7 reveal the molecular basis for ISG15 activation and E1-E2 thioester transfer.
Nat Commun, 14, 2023
|
|
6WUU
 
 | |
6WX4
 
 | |
7SOL
 
 | | Crystal Structures of the bispecific ubiquitin/FAT10 activating enzyme, Uba6 | | 分子名称: | ADENOSINE MONOPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Ubiquitin, ... | | 著者 | Olsen, S.K, Gao, F, Lv, Z. | | 登録日 | 2021-10-31 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (2.25000644 Å) | | 主引用文献 | Crystal structures reveal catalytic and regulatory mechanisms of the dual-specificity ubiquitin/FAT10 E1 enzyme Uba6.
Nat Commun, 13, 2022
|
|
4XSZ
 
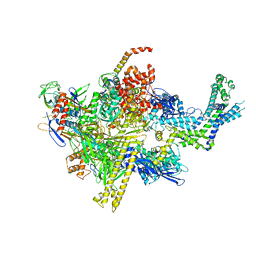 | | Crystal structure of CBR 9393 bound to Escherichia coli RNA polymerase holoenzyme | | 分子名称: | 4-[3-(4-fluorophenyl)-1H-pyrazol-4-yl]-N-[2-(piperazin-1-yl)ethyl]-2-(trifluoromethyl)aniline, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Bae, B, Darst, S.A. | | 登録日 | 2015-01-22 | | 公開日 | 2015-07-22 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.683 Å) | | 主引用文献 | CBR antimicrobials inhibit RNA polymerase via at least two bridge-helix cap-mediated effects on nucleotide addition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XSX
 
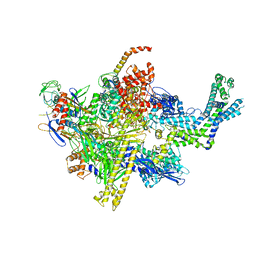 | |
4XSY
 
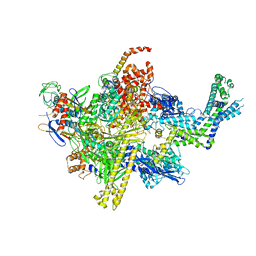 | | Crystal structure of CBR 9379 bound to Escherichia coli RNA polymerase holoenzyme | | 分子名称: | 3-{[(2,6-dichlorophenyl)carbamoyl]amino}-N-hydroxy-N'-phenyl-5-(trifluoromethyl)benzenecarboximidamide, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Bae, B, Darst, S.A. | | 登録日 | 2015-01-22 | | 公開日 | 2015-07-22 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (4.007 Å) | | 主引用文献 | CBR antimicrobials inhibit RNA polymerase via at least two bridge-helix cap-mediated effects on nucleotide addition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5XYA
 
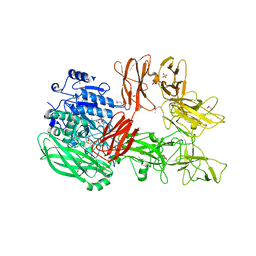 | | Crystal structure of a serine protease from Streptococcus species | | 分子名称: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CALCIUM ION, Chemokine protease C, ... | | 著者 | Jobichen, C, Sivaraman, J. | | 登録日 | 2017-07-06 | | 公開日 | 2018-08-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of ScpC, a virulence protease fromStreptococcus pyogenes, reveals the functional domains and maturation mechanism.
Biochem. J., 475, 2018
|
|
5XYR
 
 | | Crystal structure of a serine protease from Streptococcus species | | 分子名称: | CALCIUM ION, CHLORIDE ION, Chemokine protease C, ... | | 著者 | Jobichen, C, Sivaraman, J. | | 登録日 | 2017-07-10 | | 公開日 | 2018-08-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of ScpC, a virulence protease fromStreptococcus pyogenes, reveals the functional domains and maturation mechanism.
Biochem. J., 475, 2018
|
|
5XXZ
 
 | |
7EQX
 
 | | Crystal structure of an Aedes aegypti procarboxypeptidase B1 | | 分子名称: | Carboxypeptidase B, ZINC ION | | 著者 | Choong, Y.K, Gavor, E, Jobichen, C, Sivaraman, J. | | 登録日 | 2021-05-05 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structure of Aedes aegypti procarboxypeptidase B1 and its binding with Dengue virus for controlling infection.
Life Sci Alliance, 5, 2022
|
|
