7UN8
 
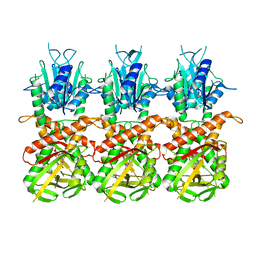 | | SfSTING with c-di-GMP single fiber | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CD-NTase-associated protein 12 | | 著者 | Morehouse, B.R, Yip, M.C.J, Keszei, A.F.A, McNamara-Bordewick, N.K, Shao, S, Kranzusch, P.J. | | 登録日 | 2022-04-09 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structure of an active bacterial TIR-STING filament complex.
Nature, 608, 2022
|
|
7UNA
 
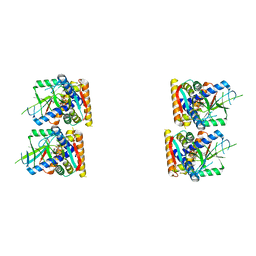 | | SfSTING with cGAMP (masked) | | 分子名称: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, CD-NTase-associated protein 12 | | 著者 | Morehouse, B.R, Yip, M.C.J, Keszei, A.F.A, McNamara-Bordewick, N.K, Shao, S, Kranzusch, P.J. | | 登録日 | 2022-04-09 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Cryo-EM structure of an active bacterial TIR-STING filament complex.
Nature, 608, 2022
|
|
7UN9
 
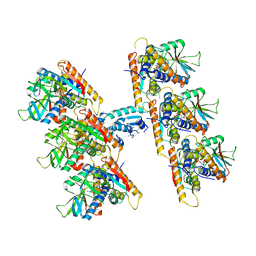 | | SfSTING with c-di-GMP double fiber | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CD-NTase-associated protein 12 | | 著者 | Morehouse, B.R, Yip, M.C.J, Keszei, A.F.A, McNamara-Bordewick, N.K, Shao, S, Kranzusch, P.J. | | 登録日 | 2022-04-09 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structure of an active bacterial TIR-STING filament complex.
Nature, 608, 2022
|
|
6WT9
 
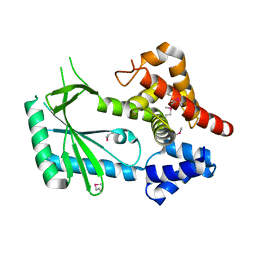 | | Structure of STING-associated CdnE c-di-GMP synthase from Capnocytophaga granulosa | | 分子名称: | NTP_transf_2 domain-containing protein | | 著者 | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | 登録日 | 2020-05-01 | | 公開日 | 2020-09-09 | | 最終更新日 | 2020-10-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
6WT6
 
 | | Structure of a metazoan TIR-STING receptor from C. gigas | | 分子名称: | Metazoan TIR-STING fusion | | 著者 | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | 登録日 | 2020-05-01 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
6WT5
 
 | | Structure of a bacterial STING receptor from Capnocytophaga granulosa | | 分子名称: | Bacterial STING | | 著者 | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | 登録日 | 2020-05-01 | | 公開日 | 2020-09-09 | | 最終更新日 | 2020-10-28 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
6WT8
 
 | | Structure of a STING-associated CdnE c-di-GMP synthase from Flavobacteriaceae sp. | | 分子名称: | STING-associated CdnE c-di-GMP synthase | | 著者 | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | 登録日 | 2020-05-01 | | 公開日 | 2020-09-09 | | 最終更新日 | 2020-10-28 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
6WT4
 
 | | Structure of a bacterial STING receptor from Flavobacteriaceae sp. in complex with 3',3'-cGAMP | | 分子名称: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Bacterial STING, SULFATE ION | | 著者 | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | 登録日 | 2020-05-01 | | 公開日 | 2020-09-09 | | 最終更新日 | 2020-10-28 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
6WT7
 
 | | Structure of a metazoan TIR-STING receptor from C. gigas in complex with 2',3'-cGAMP | | 分子名称: | Metazoan TIR-STING fusion, cGAMP | | 著者 | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | 登録日 | 2020-05-01 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
6ONM
 
 | | Crystal Structure of (+)-Limonene Synthase Complexed with 8,9-Difluorolinalyl Diphosphate | | 分子名称: | (+)-limonene synthase, (3R)-8-fluoro-7-(fluoromethyl)-3-methylocta-1,6-dien-3-yl trihydrogen diphosphate, MANGANESE (II) ION | | 著者 | Prem Kumar, R, Morehouse, B.R, Yu, Q, Oprian, D.D. | | 登録日 | 2019-04-22 | | 公開日 | 2019-09-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Direct Evidence of an Enzyme-Generated LPP Intermediate in (+)-Limonene Synthase Using a Fluorinated GPP Substrate Analog.
Acs Chem.Biol., 14, 2019
|
|
8GJX
 
 | | Structure of the human STING receptor bound to 2'3'-cUA | | 分子名称: | 2'3'-cUA, Stimulator of interferon genes protein | | 著者 | Morehouse, B.R, Li, Y, Slavik, K.M, Toyoda, H, Kranzusch, P.J. | | 登録日 | 2023-03-16 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
7R65
 
 | |
7LT1
 
 | |
6M7K
 
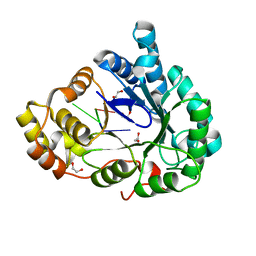 | | Structure of mouse RECON (AKR1C13) in complex with cyclic AMP-AMP-GMP (cAAG) | | 分子名称: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C13, cyclic AMP-AMP-GMP | | 著者 | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | 登録日 | 2018-08-20 | | 公開日 | 2019-02-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
8GJZ
 
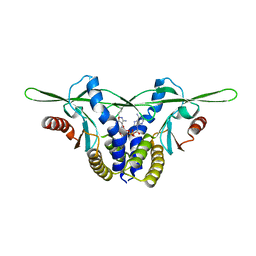 | | Structure of a STING receptor from S. pistillata Sp-STING1 bound to 2'3'-cUA | | 分子名称: | 2'3'-cUA, Stimulator of interferon genes protein | | 著者 | Li, Y, Toyoda, H, Slavik, K.M, Morehouse, B.R, Kranzusch, P.J. | | 登録日 | 2023-03-16 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8GJW
 
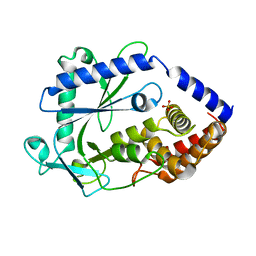 | | Structure of a cGAS-like receptor Cv-cGLR1 from C. virginica | | 分子名称: | SULFATE ION, cGAS-like receptor 1 | | 著者 | Li, Y, Morehouse, B.R, Slavik, K.M, Liu, J, Toyoda, H, Kranzusch, P.J. | | 登録日 | 2023-03-16 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8GJY
 
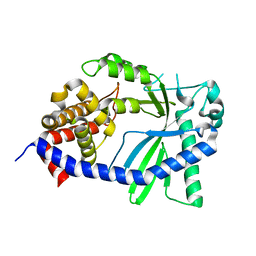 | | Structure of a cGAS-like receptor Sp-cGLR1 from S. pistillata | | 分子名称: | cGAS-like receptor 1 | | 著者 | Li, Y, Toyoda, H, Slavik, K.M, Morehouse, B.R, Kranzusch, P.J. | | 登録日 | 2023-03-16 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8EFN
 
 | | Structure of Sp-STING3 from Stylophora pistillata coral in complex with 3',3'-cGAMP | | 分子名称: | 1,2-ETHANEDIOL, 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Stimulator of interferon genes protein | | 著者 | Li, Y, Slavik, K.M, Morehouse, B.R, Mears, K, Kranzusch, P.J. | | 登録日 | 2022-09-08 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8EFM
 
 | | Structure of coral STING receptor from Stylophora pistillata in complex with 2',3'-cGAMP | | 分子名称: | SULFATE ION, Stimulator of interferon genes protein, cGAMP | | 著者 | Li, Y, Slavik, K.M, Morehouse, B.R, Mears, K, Kranzusch, P.J. | | 登録日 | 2022-09-08 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
7T26
 
 | | Structure of phage FBB1 anti-CBASS nuclease Acb1 in apo state | | 分子名称: | Acb1 | | 著者 | Hobbs, S.J, Wein, T, Lu, A, Morehouse, B.R, Schnabel, J, Sorek, R, Kranzusch, P.J. | | 登録日 | 2021-12-03 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.14 Å) | | 主引用文献 | Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity.
Nature, 605, 2022
|
|
7T28
 
 | | Structure of phage Bsp38 anti-Pycsar nuclease Apyc1 in apo state | | 分子名称: | Putative metal-dependent hydrolase, ZINC ION | | 著者 | Hobbs, S.J, Wein, T, Lu, A, Morehouse, B.R, Schnabel, J, Sorek, R, Kranzusch, P.J. | | 登録日 | 2021-12-03 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity.
Nature, 605, 2022
|
|
7T27
 
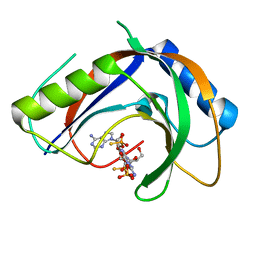 | | Structure of phage FBB1 anti-CBASS nuclease Acb1-3'3'-cGAMP complex in post reaction state | | 分子名称: | Acb1, SULFATE ION, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-2-[[[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-2-(hydroxymethyl)-4-oxidanyl-oxolan-3-yl]oxy-sulfanyl-phosphoryl]oxymethyl]-4-oxidanyl-oxolan-3-yl]oxy-sulfanyl-phosphinic acid | | 著者 | Hobbs, S.J, Wein, T, Lu, A, Morehouse, B.R, Schnabel, J, Sorek, R, Kranzusch, P.J. | | 登録日 | 2021-12-03 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity.
Nature, 605, 2022
|
|
7U2R
 
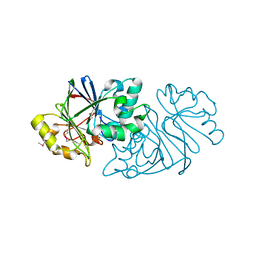 | | Structure of Paenibacillus sp. J14 Apyc1 | | 分子名称: | Apyc1, ZINC ION | | 著者 | Hobbs, S.J, Wein, T, Lu, A, Morehouse, B.R, Schnabel, J, Sorek, R, Kranzusch, P.J. | | 登録日 | 2022-02-24 | | 公開日 | 2022-04-20 | | 最終更新日 | 2022-06-01 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity.
Nature, 605, 2022
|
|
7U2S
 
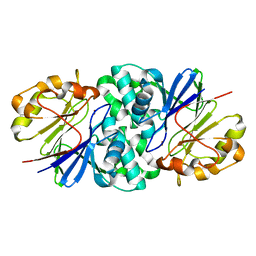 | | Structure of Paenibacillus xerothermodurans Apyc1 in the apo state | | 分子名称: | Apyc1, ZINC ION | | 著者 | Hobbs, S.J, Wein, T, Lu, A, Morehouse, B.R, Schnabel, J, Sorek, R, Kranzusch, P.J. | | 登録日 | 2022-02-24 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity.
Nature, 605, 2022
|
|
7UAW
 
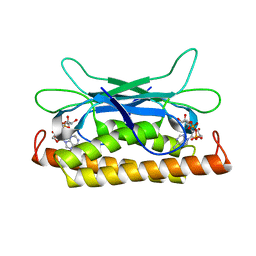 | | Structure of Clostridium botulinum prophage Tad1 in complex with 1''-2' gcADPR | | 分子名称: | (1S,3R,4R,6R,9S,11R,14R,15S,16R,18R)-4-(6-amino-9H-purin-9-yl)-9,11,15,16,18-pentahydroxy-2,5,8,10,12,17-hexaoxa-9lambda~5~,11lambda~5~-diphosphatricyclo[12.2.1.1~3,6~]octadecane-9,11-dione, ABC transporter ATPase | | 著者 | Lu, A, Leavitt, A, Yirmiya, E, Amitai, G, Garb, J, Morehouse, B.R, Hobbs, S.J, Sorek, R, Kranzusch, P.J. | | 登録日 | 2022-03-14 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Viruses inhibit TIR gcADPR signalling to overcome bacterial defence.
Nature, 611, 2022
|
|
