4BNP
 
 | | 3D structure of E. coli Isocitrate Dehydrogenase K100M mutant in complex with isocitrate and magnesium(II) | | 分子名称: | ISOCITRATE DEHYDROGENASE [NADP], ISOCITRIC ACID, MAGNESIUM ION, ... | | 著者 | Miller, S.P, Goncalves, S, Matias, P.M, Dean, A.M. | | 登録日 | 2013-05-16 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Evolution of a Transition State: Role of Lys100 in the Active Site of Isocitrate Dehydrogenase.
Chembiochem, 15, 2014
|
|
4AJB
 
 | | 3D structure of E. coli Isocitrate Dehydrogenase K100M mutant in complex with Isocitrate, magnesium(II) and thioNADP | | 分子名称: | 7-THIONICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ISOCITRIC ACID, MAGNESIUM ION, ... | | 著者 | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | 登録日 | 2012-02-16 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4AJS
 
 | | 3D structure of E. coli Isocitrate Dehydrogenase K100M mutant in complex with isocitrate, magnesium(II), Adenosine 2',5'-biphosphate and ribosylnicotinamide-5'-phosphate | | 分子名称: | ADENOSINE-2'-5'-DIPHOSPHATE, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, ISOCITRATE DEHYDROGENASE [NADP], ... | | 著者 | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | 登録日 | 2012-02-17 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4AJR
 
 | | 3D structure of E. coli Isocitrate Dehydrogenase K100M mutant in complex with alpha-ketoglutarate, magnesium(II) and NADPH - The product complex | | 分子名称: | 2-OXOGLUTARIC ACID, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, ISOCITRATE DEHYDROGENASE [NADP], ... | | 著者 | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | 登録日 | 2012-02-17 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.687 Å) | | 主引用文献 | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4AJC
 
 | | 3D structure of E. coli Isocitrate Dehydrogenase K100M mutant in complex with alpha-ketoglutarate, calcium(II) and adenine nucleotide phosphate | | 分子名称: | 2-OXOGLUTARIC ACID, ADENOSINE-2'-5'-DIPHOSPHATE, CALCIUM ION, ... | | 著者 | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | 登録日 | 2012-02-16 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4AJ3
 
 | | 3D structure of E. coli Isocitrate Dehydrogenase in complex with Isocitrate, calcium(II) and NADP - The pseudo-Michaelis complex | | 分子名称: | CALCIUM ION, ISOCITRIC ACID, NADP ISOCITRATE DEHYDROGENASE, ... | | 著者 | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | 登録日 | 2012-02-15 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4AJA
 
 | | 3D structure of E. coli Isocitrate Dehydrogenase in complex with Isocitrate, calcium(II) and thioNADP | | 分子名称: | 7-THIONICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, CALCIUM ION, ISOCITRIC ACID, ... | | 著者 | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | 登録日 | 2012-02-16 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.799 Å) | | 主引用文献 | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
1L1O
 
 | | Structure of the human Replication Protein A (RPA) trimerization core | | 分子名称: | Replication protein A 14 kDa subunit, Replication protein A 32 kDa subunit, Replication protein A 70 kDa DNA-binding subunit, ... | | 著者 | Bochkareva, E.V, Korolev, S, Lees-Miller, S.P, Bochkarev, A. | | 登録日 | 2002-02-19 | | 公開日 | 2002-06-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA.
EMBO J., 21, 2002
|
|
3SR2
 
 | | Crystal Structure of Human XLF-XRCC4 Complex | | 分子名称: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | 著者 | Hammel, M, Classen, S, Tainer, J.A. | | 登録日 | 2011-07-06 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.9708 Å) | | 主引用文献 | XRCC4 Protein Interactions with XRCC4-like Factor (XLF) Create an Extended Grooved Scaffold for DNA Ligation and Double Strand Break Repair.
J.Biol.Chem., 286, 2011
|
|
8EZB
 
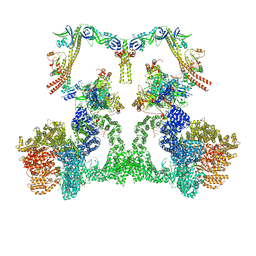 | | NHEJ Long-range complex with ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, DNA (30-MER), DNA (31-MER), ... | | 著者 | Chen, S, He, Y. | | 登録日 | 2022-10-31 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (8.9 Å) | | 主引用文献 | Cryo-EM visualization of DNA-PKcs structural intermediates in NHEJ.
Sci Adv, 9, 2023
|
|
8EZA
 
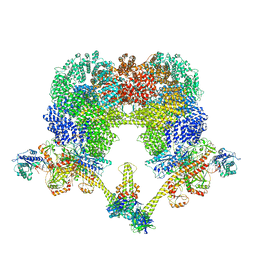 | | NHEJ Long-range complex with PAXX | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, DNA (30-MER), DNA (31-MER), ... | | 著者 | Chen, S, He, Y. | | 登録日 | 2022-10-31 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (4.39 Å) | | 主引用文献 | Cryo-EM visualization of DNA-PKcs structural intermediates in NHEJ.
Sci Adv, 9, 2023
|
|
8EZ9
 
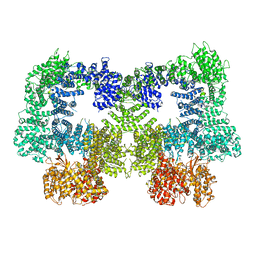 | | Dimeric complex of DNA-PKcs | | 分子名称: | DNA-dependent protein kinase catalytic subunit, unknown region of DNA-PKcs | | 著者 | Chen, S, He, Y. | | 登録日 | 2022-10-31 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (5.67 Å) | | 主引用文献 | Cryo-EM visualization of DNA-PKcs structural intermediates in NHEJ.
Sci Adv, 9, 2023
|
|
6DCX
 
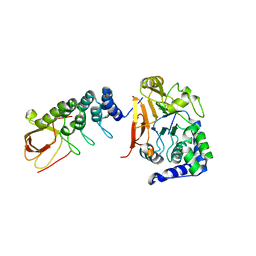 | |
7LSY
 
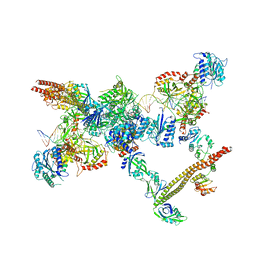 | | NHEJ Short-range synaptic complex | | 分子名称: | DNA (26-MER), DNA (5'-D(P*CP*AP*AP*TP*GP*AP*AP*AP*CP*GP*GP*AP*AP*CP*AP*GP*TP*CP*AP*G)-3'), DNA (5'-D(P*GP*TP*TP*CP*TP*TP*AP*GP*TP*AP*TP*AP*TP*A)-3'), ... | | 著者 | He, Y, Chen, S. | | 登録日 | 2021-02-18 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (8.4 Å) | | 主引用文献 | Structural basis of long-range to short-range synaptic transition in NHEJ.
Nature, 593, 2021
|
|
7LT3
 
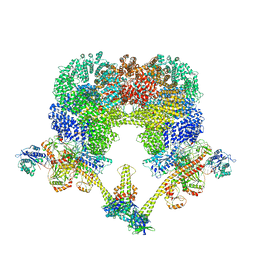 | | NHEJ Long-range synaptic complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (30-MER), DNA (31-MER), ... | | 著者 | He, Y, Chen, S. | | 登録日 | 2021-02-18 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structural basis of long-range to short-range synaptic transition in NHEJ.
Nature, 593, 2021
|
|
7MHC
 
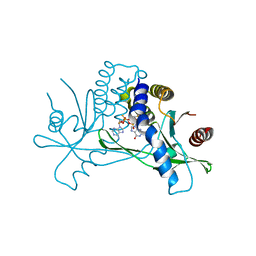 | | Structure of human STING in complex with MK-1454 | | 分子名称: | (2R,5R,7R,8S,10R,12aR,14R,15S,15aR,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-14-(6-amino-9H-purin-9-yl)-15,16-difluoro-2,10-bis(sulfanyl)octahydro-2H,10H,12H-5,8-methano-2lambda~5~,10lambda~5~-furo[3,2-l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | 著者 | Lesburg, C.A. | | 登録日 | 2021-04-15 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | A kinase-cGAS cascade to synthesize a therapeutic STING activator.
Nature, 603, 2022
|
|
