6M2W
 
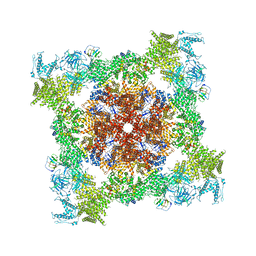 | | Structure of RyR1 (Ca2+/Caffeine/ATP/CaM1234/CHL) | | 分子名称: | 5-bromanyl-N-[4-chloranyl-2-methyl-6-(methylcarbamoyl)phenyl]-2-(3-chloranylpyridin-2-yl)pyrazole-3-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, ... | | 著者 | Ma, R, Haji-Ghassemi, O, Ma, D, Lin, L, Samurkas, A, Van Petegem, F, Yuchi, Z. | | 登録日 | 2020-03-01 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for diamide modulation of ryanodine receptor.
Nat.Chem.Biol., 16, 2020
|
|
6ADI
 
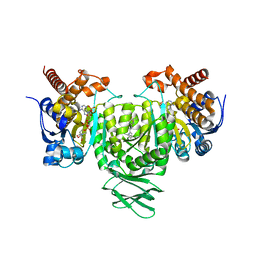 | | Crystal Structures of IDH2 R140Q in complex with AG-881 | | 分子名称: | 6-(6-chloropyridin-2-yl)-N2,N4-bis[(2R)-1,1,1-trifluoropropan-2-yl]-1,3,5-triazine-2,4-diamine, Isocitrate dehydrogenase [NADP], mitochondrial, ... | | 著者 | Ma, R, Yun, C.H. | | 登録日 | 2018-08-01 | | 公開日 | 2018-09-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.969 Å) | | 主引用文献 | Crystal structures of pan-IDH inhibitor AG-881 in complex with mutant human IDH1 and IDH2
Biochem. Biophys. Res. Commun., 503, 2018
|
|
7CF9
 
 | | Structure of RyR1 (Ca2+/CHL) | | 分子名称: | 5-bromanyl-N-[4-chloranyl-2-methyl-6-(methylcarbamoyl)phenyl]-2-(3-chloranylpyridin-2-yl)pyrazole-3-carboxamide, CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | 著者 | Ma, R, Haji-Ghassemi, O, Ma, D, Lin, L, Samurkas, A, Van Petegem, F, Yuchi, Z. | | 登録日 | 2020-06-24 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Structural basis for diamide modulation of ryanodine receptor.
Nat.Chem.Biol., 16, 2020
|
|
5GYY
 
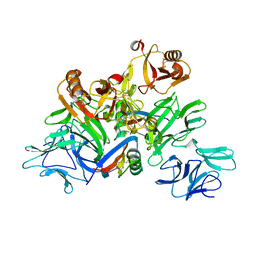 | | Plant receptor complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S-locus protein 11, S-receptor kinase SRK9 | | 著者 | Ma, R, Han, Z, Hu, Z, Lin, G, Gong, X, Zhang, H, June, N, Chai, J. | | 登録日 | 2016-09-24 | | 公開日 | 2017-09-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.351 Å) | | 主引用文献 | Plant receptor complex at 2.35 Angstroms resolution
To Be Published
|
|
7JGG
 
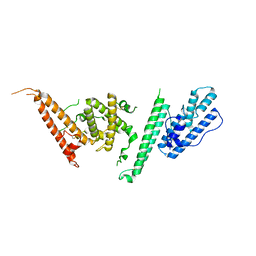 | |
7JGF
 
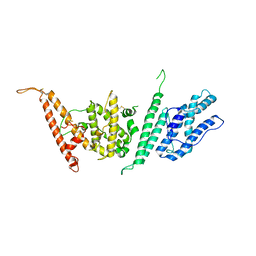 | |
7JGE
 
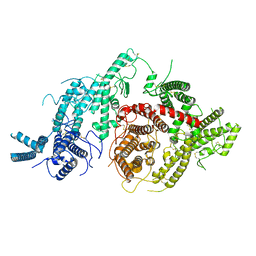 | |
7JGH
 
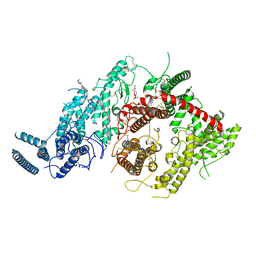 | | Cryo-EM structure of P. falciparum VAR2CSA NF54 core in complex with CSA at 3.36 A | | 分子名称: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, Erythrocyte membrane protein 1, beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid | | 著者 | Ma, R, Tolia, N.H. | | 登録日 | 2020-07-19 | | 公開日 | 2021-01-13 | | 最終更新日 | 2021-03-10 | | 実験手法 | ELECTRON MICROSCOPY (3.36 Å) | | 主引用文献 | Structural basis for placental malaria mediated by Plasmodium falciparum VAR2CSA.
Nat Microbiol, 6, 2021
|
|
7JGD
 
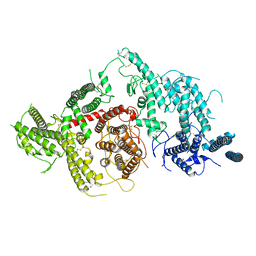 | |
6ADG
 
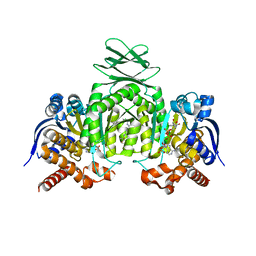 | | Crystal Structures of IDH1 R132H in complex with AG-881 | | 分子名称: | 6-(6-chloropyridin-2-yl)-N2,N4-bis[(2R)-1,1,1-trifluoropropan-2-yl]-1,3,5-triazine-2,4-diamine, Isocitrate dehydrogenase [NADP] cytoplasmic, MAGNESIUM ION, ... | | 著者 | Ma, R, Yun, C.H. | | 登録日 | 2018-08-01 | | 公開日 | 2018-09-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structures of pan-IDH inhibitor AG-881 in complex with mutant human IDH1 and IDH2
Biochem. Biophys. Res. Commun., 503, 2018
|
|
6J6L
 
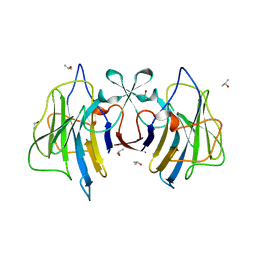 | |
7F9I
 
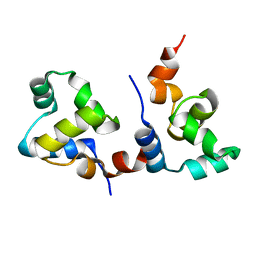 | | The apo-form structure of EnrR | | 分子名称: | EnrR repressor | | 著者 | Gan, J.H, Wang, Q.Y. | | 登録日 | 2021-07-04 | | 公開日 | 2022-05-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Xenogeneic nucleoid-associated EnrR thwarts H-NS silencing of bacterial virulence with unique DNA binding.
Nucleic Acids Res., 50, 2022
|
|
7F9H
 
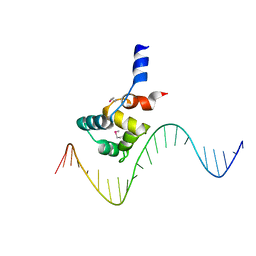 | | complex structure of EnrR-DNA | | 分子名称: | EnrR repressor, target DNA | | 著者 | Gan, J.H, Wang, Q.Y. | | 登録日 | 2021-07-04 | | 公開日 | 2022-05-11 | | 最終更新日 | 2022-05-18 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Xenogeneic nucleoid-associated EnrR thwarts H-NS silencing of bacterial virulence with unique DNA binding.
Nucleic Acids Res., 50, 2022
|
|
6L0X
 
 | | The First Tudor Domain of PHF20L1 | | 分子名称: | CITRIC ACID, GLYCEROL, PHD finger protein 20-like protein 1 | | 著者 | Lv, M.Q, Gao, J. | | 登録日 | 2019-09-27 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1P
 
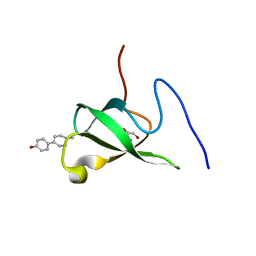 | | Crystal structure of PHF20L1 in complex with Hit 1 | | 分子名称: | 4-(1-methyl-3,6-dihydro-2H-pyridin-4-yl)phenol, GLYCEROL, PHD finger protein 20-like protein 1, ... | | 著者 | Lv, M.Q, Gao, J. | | 登録日 | 2019-09-29 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.231 Å) | | 主引用文献 | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1C
 
 | |
6L1I
 
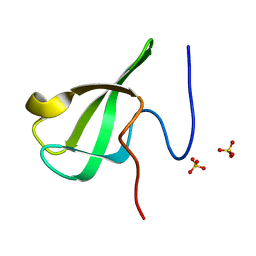 | |
6L10
 
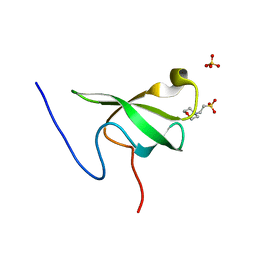 | | PHF20L1 Tudor1 - MES | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PHD finger protein 20-like protein 1, SULFATE ION | | 著者 | Lv, M.Q, Gao, J. | | 登録日 | 2019-09-27 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
8HHT
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Hit-1 | | 分子名称: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(1,3-thiazol-2-ylmethylamino)butan-2-yl]benzamide | | 著者 | Zeng, R, Xie, L.W, Huang, C, Wang, K, Liu, Y.Z, Yang, S.Y, Lei, J. | | 登録日 | 2022-11-17 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A new generation M pro inhibitor with potent activity against SARS-CoV-2 Omicron variants.
Signal Transduct Target Ther, 8, 2023
|
|
8HHU
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with SY110 | | 分子名称: | (1~{R})-3,3-bis(fluoranyl)-~{N}-[(2~{R})-3-methoxy-1-oxidanylidene-1-[[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(1,3-thiazol-2-ylmethylamino)butan-2-yl]amino]propan-2-yl]cyclohexane-1-carboxamide, 3C-like proteinase nsp5 | | 著者 | Zeng, R, Xie, L.W, Huang, C, Wang, K, Liu, Y.Z, Yang, S.Y, Lei, J. | | 登録日 | 2022-11-17 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.258 Å) | | 主引用文献 | A new generation M pro inhibitor with potent activity against SARS-CoV-2 Omicron variants.
Signal Transduct Target Ther, 8, 2023
|
|
8I30
 
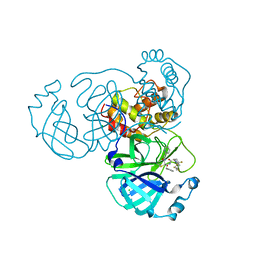 | | Crystal structure of the SARS-CoV-2 main protease in complex with 32j | | 分子名称: | (2~{R})-1-[4,4-bis(fluoranyl)cyclohexyl]carbonyl-4,4-bis(fluoranyl)-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]pyrrolidine-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | 著者 | Zeng, R, Huang, C, Xie, L.W, Wang, K, Liu, Y.Z, Yang, S.Y, Lei, J. | | 登録日 | 2023-01-16 | | 公開日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery and structure-activity relationship studies of novel alpha-ketoamide derivatives targeting the SARS-CoV-2 main protease.
Eur.J.Med.Chem., 259, 2023
|
|
4KXF
 
 | |
5H1U
 
 | | Complex structure of TRIM24 PHD-bromodomain and inhibitor 2 | | 分子名称: | 2-amino-1,3-benzothiazole-6-carboxamide, DIMETHYL SULFOXIDE, Transcription intermediary factor 1-alpha, ... | | 著者 | Liu, J. | | 登録日 | 2016-10-11 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | The polar warhead of a TRIM24 bromodomain inhibitor rearranges a water-mediated interaction network
FEBS J., 284, 2017
|
|
5H1T
 
 | | Complex structure of TRIM24 PHD-bromodomain and inhibitor 1 | | 分子名称: | DIMETHYL SULFOXIDE, Transcription intermediary factor 1-alpha, ZINC ION, ... | | 著者 | Liu, J. | | 登録日 | 2016-10-11 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.951 Å) | | 主引用文献 | The polar warhead of a TRIM24 bromodomain inhibitor rearranges a water-mediated interaction network
FEBS J., 284, 2017
|
|
5H1D
 
 | | Crystal structure of C-terminal of RhoGDI2 | | 分子名称: | Rho GDP-dissociation inhibitor 2 | | 著者 | Liu, J. | | 登録日 | 2016-10-08 | | 公開日 | 2016-10-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.494 Å) | | 主引用文献 | NMR characterization of weak interactions between RhoGDI2 and fragment screening hits.
Biochim. Biophys. Acta, 1861, 2017
|
|
