8V8E
 
 | | Room-temperature X-ray structure of SARS-CoV-2 main protease catalytic domain (residues 1-199-6H) in complex with ensitrelvir (ESV) | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, peptide | | 著者 | Kovalevsky, A, Coates, L. | | 登録日 | 2023-12-05 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Visualizing the Active Site Oxyanion Loop Transition Upon Ensitrelvir Binding and Transient Dimerization of SARS-CoV-2 Main Protease.
J.Mol.Biol., 436, 2024
|
|
8V7W
 
 | |
8V8G
 
 | | Room-temperature X-ray structure of SARS-CoV-2 main protease catalytic domain (residues 1-196) in complex with ensitrelvir (ESV) | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Kovalevsky, A, Coates, L. | | 登録日 | 2023-12-05 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Visualizing the Active Site Oxyanion Loop Transition Upon Ensitrelvir Binding and Transient Dimerization of SARS-CoV-2 Main Protease.
J.Mol.Biol., 436, 2024
|
|
8V7T
 
 | |
8GFO
 
 | |
8GFR
 
 | | Room temperature X-ray structure of truncated SARS-CoV-2 main protease C145A mutant, residues 1-304, in complex with NBH2 | | 分子名称: | (1R,2S,5S)-N-{(1S)-1-cyano-2-[(3S)-2-oxopyrrolidin-3-yl]ethyl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Kovalevsky, A, Coates, L. | | 登録日 | 2023-03-08 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Contribution of the catalytic dyad of SARS-CoV-2 main protease to binding covalent and noncovalent inhibitors.
J.Biol.Chem., 299, 2023
|
|
8GFU
 
 | |
8GFK
 
 | |
8GFN
 
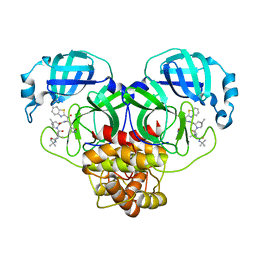 | | Room temperature X-ray structure of truncated SARS-CoV-2 main protease C145A mutant, residues 1-304, in complex with BBH1 | | 分子名称: | (1R,2S,5S)-N-{(2S)-1-(1,3-benzothiazol-2-yl)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Kovalevsky, A, Coates, L. | | 登録日 | 2023-03-08 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of the catalytic dyad of SARS-CoV-2 main protease to binding covalent and noncovalent inhibitors.
J.Biol.Chem., 299, 2023
|
|
9CMS
 
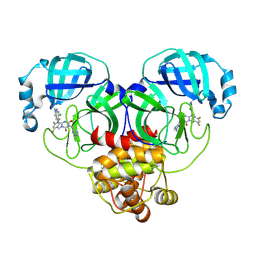 | | Room-temperature X-ray structure of SARS-CoV-2 main protease drug resistant mutant (E166V) in complex with ensitrelvir (ESV) | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Kovalevsky, A, Coates, L, Gerlits, O. | | 登録日 | 2024-07-15 | | 公開日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Effects of SARS-CoV-2 Main Protease Mutations at Positions L50, E166, and L167 Rendering Resistance to Covalent and Noncovalent Inhibitors.
J.Med.Chem., 2024
|
|
9CMN
 
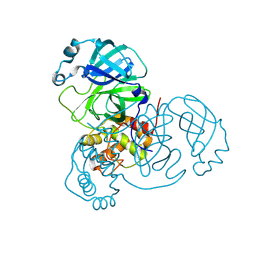 | |
9CMJ
 
 | |
9CMU
 
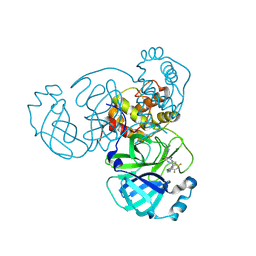 | | Room-temperature X-ray structure of SARS-CoV-2 main protease drug resistant mutant (L50F, E166V) in complex with ensitrelvir (ESV) | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Kovalevsky, A, Coates, L, Gerlits, O. | | 登録日 | 2024-07-15 | | 公開日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Effects of SARS-CoV-2 Main Protease Mutations at Positions L50, E166, and L167 Rendering Resistance to Covalent and Noncovalent Inhibitors.
J.Med.Chem., 2024
|
|
6BBS
 
 | | Joint X-ray/neutron structure of human carbonic anhydrase II in complex with brinzolamide | | 分子名称: | (+)-4-ETHYLAMINO-3,4-DIHYDRO-2-(METHOXY)PROPYL-2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, Carbonic anhydrase 2, ZINC ION | | 著者 | Kovalevsky, A, Aggarwal, M, McKenna, R. | | 登録日 | 2017-10-19 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | 主引用文献 | "To Be or Not to Be" Protonated: Atomic Details of Human Carbonic Anhydrase-Clinical Drug Complexes by Neutron Crystallography and Simulation.
Structure, 26, 2018
|
|
6BCC
 
 | | Joint X-ray/neutron structure of human carbonic anhydrase II in complex with ethoxzolamide | | 分子名称: | 6-ethoxy-1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | 著者 | Kovalevsky, A, McKenna, R, Aggarwal, M. | | 登録日 | 2017-10-20 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | 主引用文献 | "To Be or Not to Be" Protonated: Atomic Details of Human Carbonic Anhydrase-Clinical Drug Complexes by Neutron Crystallography and Simulation.
Structure, 26, 2018
|
|
6BC9
 
 | | Joint X-ray/neutron structure of human carbonic anhydrase II in complex with dorzolamide | | 分子名称: | (4S-TRANS)-4-(ETHYLAMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO(2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, Carbonic anhydrase 2, ZINC ION | | 著者 | Kovalevsky, A, McKenna, R, Aggarwal, M. | | 登録日 | 2017-10-20 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | 主引用文献 | "To Be or Not to Be" Protonated: Atomic Details of Human Carbonic Anhydrase-Clinical Drug Complexes by Neutron Crystallography and Simulation.
Structure, 26, 2018
|
|
5WEY
 
 | | Joint X-ray/neutron structure of Concanavalin A with alpha1-2 D-mannobiose | | 分子名称: | CALCIUM ION, Concanavalin-A, MANGANESE (II) ION, ... | | 著者 | Kovalevsky, A, Gerlits, O.O, Woods, R.J. | | 登録日 | 2017-07-11 | | 公開日 | 2017-09-13 | | 最終更新日 | 2023-10-04 | | 実験手法 | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | 主引用文献 | Mannobiose Binding Induces Changes in Hydrogen Bonding and Protonation States of Acidic Residues in Concanavalin A As Revealed by Neutron Crystallography.
Biochemistry, 56, 2017
|
|
7JUN
 
 | |
8E4R
 
 | |
8E4J
 
 | |
6U34
 
 | | Binary complex of native hAChE with oxime reactivator RS194B | | 分子名称: | (2E)-N-[2-(azepan-1-yl)ethyl]-2-(hydroxyimino)acetamide, Acetylcholinesterase, GLYCEROL, ... | | 著者 | Kovalevsky, A, Gerlits, O, Radic, Z. | | 登録日 | 2019-08-21 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Rational design, synthesis, and evaluation of uncharged, "smart" bis-oxime antidotes of organophosphate-inhibited human acetylcholinesterase.
J.Biol.Chem., 295, 2020
|
|
6U37
 
 | | Structure of VX-phosphonylated hAChE in complex with oxime reactivator RS194B | | 分子名称: | (2E)-N-[2-(azepan-1-yl)ethyl]-2-(hydroxyimino)acetamide, Acetylcholinesterase, GLYCEROL, ... | | 著者 | Kovalevsky, A, Gerlits, O, Radic, Z. | | 登録日 | 2019-08-21 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Rational design, synthesis, and evaluation of uncharged, "smart" bis-oxime antidotes of organophosphate-inhibited human acetylcholinesterase.
J.Biol.Chem., 295, 2020
|
|
6U3P
 
 | | Binary complex of native hAChE with oxime reactivator LG-703 | | 分子名称: | (2E,2'E)-N,N'-[1,4-diazepane-1,4-diyldi(ethane-2,1-diyl)]bis[2-(hydroxyimino)acetamide], Acetylcholinesterase, DIMETHYL SULFOXIDE | | 著者 | Kovalevsky, A, Gerlits, O, Radic, Z. | | 登録日 | 2019-08-22 | | 公開日 | 2020-02-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Rational design, synthesis, and evaluation of uncharged, "smart" bis-oxime antidotes of organophosphate-inhibited human acetylcholinesterase.
J.Biol.Chem., 295, 2020
|
|
7LTJ
 
 | | Room-temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with a non-covalent inhibitor Mcule-5948770040 | | 分子名称: | 3C-like proteinase, 6-[4-(3,4-dichlorophenyl)piperazin-1-yl]carbonyl-1~{H}-pyrimidine-2,4-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-02-19 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | High-Throughput Virtual Screening and Validation of a SARS-CoV-2 Main Protease Noncovalent Inhibitor.
J.Chem.Inf.Model., 62, 2022
|
|
7UKK
 
 | |
