5G4F
 
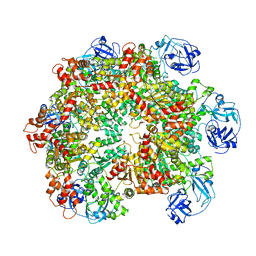 | | Structure of the ADP-bound VAT complex | | 分子名称: | VCP-LIKE ATPASE | | 著者 | Huang, R, Ripstein, Z.A, Augustyniak, R, Lazniewski, M, Ginalski, K, Kay, L.E, Rubinstein, J.L. | | 登録日 | 2016-05-12 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Unfolding the Mechanism of the Aaa+ Unfoldase Vat by a Combined Cryo-Em, Solution NMR Study.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5G4G
 
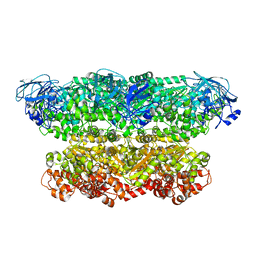 | | Structure of the ATPgS-bound VAT complex | | 分子名称: | VCP-LIKE ATPASE | | 著者 | Huang, R, Ripstein, Z.A, Augustyniak, R, Lazniewski, M, Ginalski, K, Kay, L.E, Rubinstein, J.L. | | 登録日 | 2016-05-12 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.8 Å) | | 主引用文献 | Unfolding the Mechanism of the Aaa+ Unfoldase Vat by a Combined Cryo-Em, Solution NMR Study.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4N9B
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | 分子名称: | 1-methyl-N-(pyridin-3-yl)-1H-pyrazole-5-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | 著者 | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhai, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | 登録日 | 2013-10-20 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.859 Å) | | 主引用文献 | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N9C
 
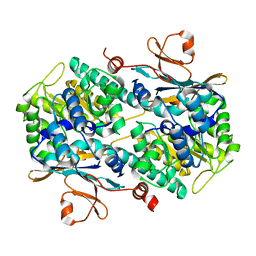 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | 分子名称: | 5-nitro-1H-benzimidazole, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | 著者 | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | 登録日 | 2013-10-20 | | 公開日 | 2014-02-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.751 Å) | | 主引用文献 | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N9E
 
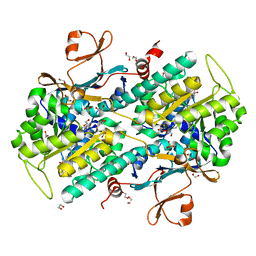 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | 分子名称: | 1,2-ETHANEDIOL, 1-[(1-benzoylpiperidin-4-yl)methyl]-N-(pyridin-3-yl)-1H-benzimidazole-5-carboxamide, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | 著者 | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | 登録日 | 2013-10-20 | | 公開日 | 2014-02-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N9D
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | 分子名称: | 1,2-ETHANEDIOL, 4-({[(4-tert-butylphenyl)sulfonyl]amino}methyl)-N-(pyridin-3-yl)benzamide, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | 登録日 | 2013-10-20 | | 公開日 | 2014-02-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
8W5K
 
 | | Cryo-EM structure of the yeast TOM core complex crosslinked by BS3 (from TOM-TIM23 complex) | | 分子名称: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | 著者 | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | 登録日 | 2023-08-27 | | 公開日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
8W5J
 
 | | Cryo-EM structure of the yeast TOM core complex (from TOM-TIM23 complex) | | 分子名称: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | 著者 | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | 登録日 | 2023-08-27 | | 公開日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
8SHB
 
 | | Crystal Structure of PRMT3 with Compound YD1-208 | | 分子名称: | 5'-S-[3-(N'-phenylcarbamimidamido)propyl]-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | 著者 | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2023-04-13 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Crystal Structure of PRMT3 with Compound YD1-208
To be published
|
|
8SIH
 
 | | Crystal Structure of PRMT4 with Compound YD1-289 | | 分子名称: | 5'-{[2-(benzylcarbamamido)ethyl][3-(N'-cyclopentylcarbamimidamido)propyl]amino}-5'-deoxyadenosine, CALCIUM ION, Histone-arginine methyltransferase CARM1 | | 著者 | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2023-04-16 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal Structure of PRMT4 with Compound YD1-289
To be published
|
|
8SIG
 
 | | Crystal Structure of PRMT4 with Compound YD1-288 | | 分子名称: | 5'-{(3-aminopropyl)[2-(benzylcarbamamido)ethyl]amino}-5'-deoxyadenosine, Histone-arginine methyltransferase CARM1, SODIUM ION, ... | | 著者 | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2023-04-16 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Crystal Structure of PRMT4 with Compound YD1-288
To be published
|
|
8SHR
 
 | | Crystal Structure of PRMT3 with Compound YD1-214 | | 分子名称: | 5'-S-[2-(phenylcarbamamido)ethyl]-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | 著者 | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2023-04-14 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Crystal Structure of PRMT3 with Compound YD1-214
To be published
|
|
8SIO
 
 | | Crystal structure of PRMT3 with YD1-66 | | 分子名称: | 5'-S-{3-[N'-(4'-chloro[1,1'-biphenyl]-3-yl)carbamimidamido]propyl}-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | 著者 | Song, X, Dong, A, Arrowsmith, C.H, Edwards, A.M, Deng, Y, Huang, R, Min, J. | | 登録日 | 2023-04-16 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of PRMT3 with YD1-66
To be published
|
|
6DTN
 
 | |
8UYJ
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | 分子名称: | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-13 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (7.3 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYS
 
 | | SARS-CoV-2 5' proximal stem-loop 5 | | 分子名称: | SARS-CoV-2 RNA SL5 domain. | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-14 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYM
 
 | | MERS 5' proximal stem-loop 5, conformation 3 | | 分子名称: | MERS 5' proximal stem-loop 5 | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-13 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYK
 
 | | MERS 5' proximal stem-loop 5, conformation 1 | | 分子名称: | MERS 5' proximal stem-loop 5 | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-13 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYL
 
 | | MERS 5' proximal stem-loop 5, conformation 2 | | 分子名称: | MERS 5' proximal stem-loop 5 | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-13 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYG
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 2 | | 分子名称: | RNA (135-MER) | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-13 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYE
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 1 | | 分子名称: | BtCoV-HKU5 5' proximal stem-loop 5 | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-13 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (5.9 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYP
 
 | | SARS-CoV-1 5' proximal stem-loop 5 | | 分子名称: | SARS-CoV-1 5' proximal stem-loop 5 | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-13 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (7.1 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6PVS
 
 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with inhibitor LL320 | | 分子名称: | 9-(5-{[(3R)-3-amino-3-carboxypropyl][3-(3-carbamoylphenyl)prop-2-yn-1-yl]amino}-5-deoxy-alpha-D-lyxofuranosyl)-9H-purin-6-amine, NNMT protein | | 著者 | Noinaj, N, Huang, R, Chen, D, Yadav, R. | | 登録日 | 2019-07-21 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.575 Å) | | 主引用文献 | Novel Propargyl-Linked Bisubstrate Analogues as Tight-Binding Inhibitors for NicotinamideN-Methyltransferase.
J.Med.Chem., 62, 2019
|
|
6PVE
 
 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with inhibitor LL319 | | 分子名称: | 9-(5-{[(3S)-3-amino-3-carboxypropyl][3-(3-carbamoylphenyl)propyl]amino}-5-deoxy-alpha-D-ribofuranosyl)-9H-purin-6-amine, NNMT protein | | 著者 | Noinaj, N, Huang, R, Chen, D, Yadav, R. | | 登録日 | 2019-07-20 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Novel Propargyl-Linked Bisubstrate Analogues as Tight-Binding Inhibitors for NicotinamideN-Methyltransferase.
J.Med.Chem., 62, 2019
|
|
7MT0
 
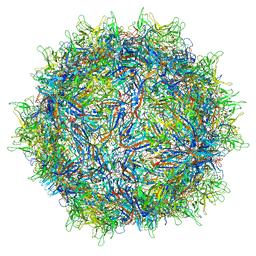 | | Structure of the adeno-associated virus 9 capsid at pH 7.4 | | 分子名称: | Capsid protein VP1 | | 著者 | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | 登録日 | 2021-05-12 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.82 Å) | | 主引用文献 | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
