4YFS
 
 | |
4QEX
 
 | | Crystal structure of PfEBA-175 RII in complex with a Fab fragment from inhibitory antibody R217 | | 分子名称: | Antibody Heavy Chain, Antibody Light Chain, Erythrocyte-binding antigen-175 | | 著者 | Chen, E, Paing, M.M, Salinas, N, Sim, B.K, Tolia, N.H. | | 登録日 | 2014-05-19 | | 公開日 | 2014-06-04 | | 実験手法 | X-RAY DIFFRACTION (4.5 Å) | | 主引用文献 | Structural and Functional Basis for Inhibition of Erythrocyte Invasion by Antibodies that Target Plasmodium falciparum EBA-175.
Plos Pathog., 9, 2013
|
|
5F3J
 
 | |
2ESN
 
 | | The crystal structure of probable transcriptional regulator PA0477 from Pseudomonas aeruginosa | | 分子名称: | probable transcriptional regulator | | 著者 | Lunin, V.V, Chang, C, Skarina, T, Gorodischenskaya, E, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-10-26 | | 公開日 | 2005-11-01 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The crystal structure of putative transcriptional regulator Pa0477 from Pseudomonas aeruginosa
To be Published
|
|
2FDO
 
 | | Crystal Structure of the Conserved Protein of Unknown Function AF2331 from Archaeoglobus fulgidus DSM 4304 Reveals a New Type of Alpha/Beta Fold | | 分子名称: | Hypothetical protein AF2331 | | 著者 | Wang, S, Kirillova, O, Chruszcz, M, Cymborowski, M.T, Skarina, T, Gorodichtchenskaia, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-12-14 | | 公開日 | 2006-01-31 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The crystal structure of the AF2331 protein from Archaeoglobus fulgidus DSM 4304 forms an unusual interdigitated dimer with a new type of alpha + beta fold.
Protein Sci., 18, 2009
|
|
2GFQ
 
 | | Structure of Protein of Unknown Function PH0006 from Pyrococcus horikoshii | | 分子名称: | MAGNESIUM ION, SULFATE ION, UPF0204 protein PH0006 | | 著者 | Cuff, M.E, Skarina, T, Gorodichtchenskaia, E, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-03-22 | | 公開日 | 2006-04-25 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure of hypothetical protein ph0006 from Pyrococcus horikoshii
To be Published
|
|
1WCO
 
 | | The solution structure of the nisin-lipid II complex | | 分子名称: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, ALA-FGA-LYS-DAL-DAL PEPTIDE, ... | | 著者 | Hsu, S.-T.D, Breukink, E, Tischenko, E, Lutters, M.A.G, de Kruijff, B, Kaptein, R, Bonvin, A.M.J.J, van Nuland, N.A.J. | | 登録日 | 2004-11-19 | | 公開日 | 2005-03-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics.
Nat. Struct. Mol. Biol., 11, 2004
|
|
1XEB
 
 | | Crystal Structure of an Acyl-CoA N-acyltransferase from Pseudomonas aeruginosa | | 分子名称: | hypothetical protein PA0115 | | 著者 | Bertero, M.G, Walker, J.R, Skarina, T, Gorodichtchenskaia, E, Joachimiak, A, Edwards, A.E, Savchenko, A, Strynadka, N, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2004-09-09 | | 公開日 | 2004-10-26 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | The crystal structure of an Acyl-CoA N-acyltransferase from Pseudomonas aeruginosa
To be Published
|
|
1Y0N
 
 | | Structure of Protein of Unknown Function PA3463 from Pseudomonas aeruginosa PAO1 | | 分子名称: | GLYCEROL, Hypothetical UPF0270 protein PA3463 | | 著者 | Binkowski, T.A, Edwards, A, Savchenko, A, Skarina, T, Gorodichtchenskaia, E, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2004-11-15 | | 公開日 | 2004-12-21 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Hypothetical protein PA3463 from Pseudomonas aeruginosa strain PAO1
To be Published
|
|
2AZP
 
 | | Crystal Structure of PA1268 Solved by Sulfur SAD | | 分子名称: | hypothetical protein PA1268 | | 著者 | Liu, Y, Gorodichtchenskaia, E, Skarina, T, Yang, C, Joachimiak, A, Edwards, A, Pai, E.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-09-12 | | 公開日 | 2005-12-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Crystal Structure of PA1268 Solved by Sulfur SAD
To be Published
|
|
5ZR1
 
 | | Saccharomyces Cerevisiae Origin Recognition Complex Bound to a 72-bp Origin DNA containing ACS and B1 element | | 分子名称: | 72bp-oring DNA, ACS305, A-rich, ... | | 著者 | Li, N, Lam, W.H, Zhai, Y, Cheng, J, Cheng, E, Zhao, Y, Gao, N, Tye, B.K. | | 登録日 | 2018-04-21 | | 公開日 | 2018-07-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of the origin recognition complex bound to DNA replication origin.
Nature, 559, 2018
|
|
2O9X
 
 | | Crystal Structure Of A Putative Redox Enzyme Maturation Protein From Archaeoglobus Fulgidus | | 分子名称: | Reductase, assembly protein | | 著者 | Kirillova, O, Chruszcz, M, Skarina, T, Gorodichtchenskaia, E, Cymborowski, M, Shumilin, I, Savchenko, A, Edwards, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-12-14 | | 公開日 | 2007-01-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | An extremely SAD case: structure of a putative redox-enzyme maturation protein from Archaeoglobus fulgidus at 3.4 A resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1K1R
 
 | | HETERODUPLEX OF CHIRALLY PURE R-METHYLPHOSPHONATE/DNA DUPLEX | | 分子名称: | 5'-D(*CP*(CMR)P*(RMP)P*(RMP)P*(RMP)P*(CMR)P*(RMP))-3', 5'-D(*TP*GP*TP*TP*TP*GP*GP*C)-3' | | 著者 | Thiviyanathan, V, Vyazovkina, K.V, Gozansky, E.K, Bichenkova, E, Abramova, T.V, Luxon, B.A, Lebedev, A.V, Gorenstein, D.G. | | 登録日 | 2001-09-25 | | 公開日 | 2002-06-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of hybrid backbone methylphosphonate DNA heteroduplexes: effect of R and S stereochemistry.
Biochemistry, 41, 2002
|
|
5XF8
 
 | | Cryo-EM structure of the Cdt1-MCM2-7 complex in AMPPNP state | | 分子名称: | Cell division cycle protein CDT1, DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, ... | | 著者 | Zhai, Y, Cheng, E, Wu, H, Li, N, Yung, P.Y, Gao, N, Tye, B.K. | | 登録日 | 2017-04-09 | | 公開日 | 2017-05-03 | | 実験手法 | ELECTRON MICROSCOPY (7.1 Å) | | 主引用文献 | Open-ringed structure of the Cdt1-Mcm2-7 complex as a precursor of the MCM double hexamer
Nat. Struct. Mol. Biol., 24, 2017
|
|
1K1H
 
 | | HETERODUPLEX OF CHIRALLY PURE METHYLPHOSPHONATE/DNA DUPLEX | | 分子名称: | 5'-D(*CP*(CMR)P*(RMP)P*(RMP)P*(SMP)P*(CMR)P*(RMP))-3', 5'-D(*TP*GP*TP*TP*TP*GP*GP*C)-3' | | 著者 | Thiviyanathan, V, Vyazovkina, K.V, Gozansky, E.K, Bichenchova, E, Abramova, T.V, Luxon, B.A, Lebedev, A.V, Gorenstein, D.G. | | 登録日 | 2001-09-25 | | 公開日 | 2002-06-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of hybrid backbone methylphosphonate DNA heteroduplexes: effect of R and S stereochemistry.
Biochemistry, 41, 2002
|
|
2LKD
 
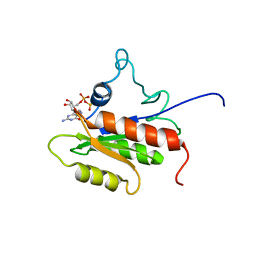 | | IF2-G2 GDP complex | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Translation initiation factor IF-2 | | 著者 | Wienk, H, Tishchenko, E, Belardinelli, R, Tomaselli, S, Dongre, R, Spurio, R, Folkers, G.E, Gualerzi, C.O, Boelens, R. | | 登録日 | 2011-10-10 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Dynamics of Bacterial Translation Initiation Factor IF2.
J.Biol.Chem., 287, 2012
|
|
2LKC
 
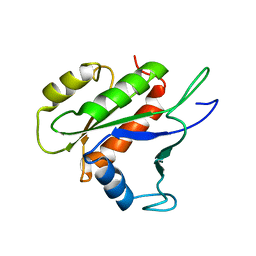 | | Free B.st IF2-G2 | | 分子名称: | Translation initiation factor IF-2 | | 著者 | Wienk, H, Tishchenko, E, Belardinelli, R, Tomaselli, S, Dongre, R, Spurio, R, Folkers, G.E, Gualerzi, C.O, Boelens, R. | | 登録日 | 2011-10-10 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Dynamics of Bacterial Translation Initiation Factor IF2.
J.Biol.Chem., 287, 2012
|
|
2KEJ
 
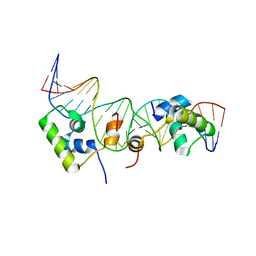 | | Solution structure of a dimer of LAC repressor DNA-binding domain complexed to its natural operator O2 | | 分子名称: | DNA (5'-D(*GP*AP*AP*AP*TP*GP*TP*GP*AP*GP*CP*GP*AP*GP*TP*AP*AP*CP*AP*AP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*TP*TP*GP*TP*TP*AP*CP*TP*CP*GP*CP*TP*CP*AP*CP*AP*TP*TP*TP*C)-3'), Lactose operon repressor | | 著者 | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | 登録日 | 2009-01-30 | | 公開日 | 2009-05-19 | | 最終更新日 | 2021-10-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
2KEI
 
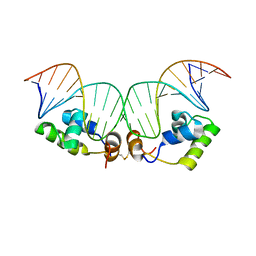 | | Refined Solution Structure of a Dimer of LAC repressor DNA-Binding domain complexed to its natural operator O1 | | 分子名称: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*GP*AP*TP*AP*AP*CP*AP*AP*TP*TP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*TP*GP*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3'), Lactose operon repressor | | 著者 | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | 登録日 | 2009-01-30 | | 公開日 | 2009-05-19 | | 最終更新日 | 2021-10-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
2KEK
 
 | | Solution structure of a dimer of LAC repressor DNA-binding domain complexed to its natural operator O3 | | 分子名称: | DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*GP*AP*GP*CP*GP*CP*AP*AP*CP*GP*CP*AP*AP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*TP*TP*GP*CP*GP*TP*TP*GP*CP*GP*CP*TP*CP*AP*CP*TP*GP*CP*CP*G)-3'), Lactose operon repressor | | 著者 | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | 登録日 | 2009-01-30 | | 公開日 | 2009-05-19 | | 最終更新日 | 2021-10-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
3JCE
 
 | | Structure of Escherichia coli EF4 in pretranslocational ribosomes (Pre EF4) | | 分子名称: | 16S ribosomal RNA, 23 ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Zhang, D, Yan, K, Liu, G, Song, G, Luo, J, Shi, Y, Cheng, E, Wu, S, Jiang, T, Low, J, Gao, N, Qin, Y. | | 登録日 | 2015-12-01 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nat. Struct. Mol. Biol., 23, 2016
|
|
3JCD
 
 | | Structure of Escherichia coli EF4 in posttranslocational ribosomes (Post EF4) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Zhang, D, Yan, K, Liu, G, Song, G, Luo, J, Shi, Y, Cheng, E, Wu, S, Jiang, T, Low, J, Gao, N, Qin, Y. | | 登録日 | 2015-12-01 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nat. Struct. Mol. Biol., 23, 2016
|
|
7MRU
 
 | | Crystal structure of S62A MIF2 mutant | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-dopachrome decarboxylase | | 著者 | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | 登録日 | 2021-05-09 | | 公開日 | 2021-08-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
7MW7
 
 | | Crystal structure of P1G mutant of D-dopachrome tautomerase | | 分子名称: | D-dopachrome decarboxylase, SODIUM ION, SULFATE ION | | 著者 | Manjula, R, Murphy, E.L, Murphy, J.W, Lolis, E. | | 登録日 | 2021-05-15 | | 公開日 | 2021-08-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
7MSE
 
 | | High-resolution crystal structure of hMIF2 with tartrate at the active site | | 分子名称: | D-dopachrome decarboxylase, L(+)-TARTARIC ACID | | 著者 | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | 登録日 | 2021-05-11 | | 公開日 | 2021-08-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
