8J9I
 
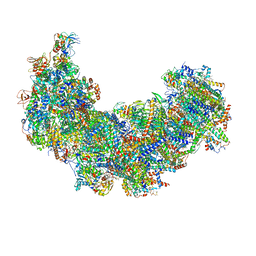 | | Cryo-EM structure of Euglena gracilis complex I, turnover state | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | 著者 | Wu, M.C, He, Z.X, Tian, H.T, Hu, Y.Q, Han, F.Z, Zhou, L. | | 登録日 | 2023-05-03 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (2.87 Å) | | 主引用文献 | Euglena's atypical respiratory chain adapts to the discoidal cristae and flexible metabolism.
Nat Commun, 15, 2024
|
|
8J9J
 
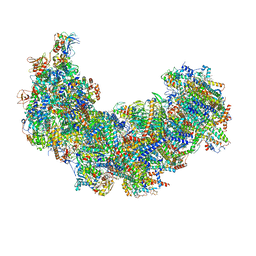 | | Cryo-EM structure of Euglena gracilis complex I, NADH state | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | 著者 | Wu, M.C, He, Z.X, Tian, H.T, Hu, Y.Q, Han, F.Z, Zhou, L. | | 登録日 | 2023-05-03 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Euglena's atypical respiratory chain adapts to the discoidal cristae and flexible metabolism.
Nat Commun, 15, 2024
|
|
6IOE
 
 | |
6INO
 
 | |
3UUE
 
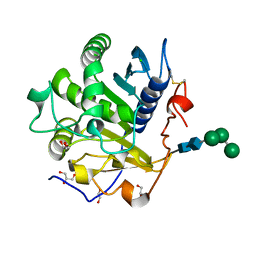 | | Crystal structure of mono- and diacylglycerol lipase from Malassezia globosa | | 分子名称: | CHLORIDE ION, GLYCEROL, LIP1, ... | | 著者 | Xu, T, Xu, J, Hou, S, Liu, J. | | 登録日 | 2011-11-28 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal structure of a mono- and diacylglycerol lipase from Malassezia globosa reveals a novel lid conformation and insights into the substrate specificity.
J.Struct.Biol., 178, 2012
|
|
6KBD
 
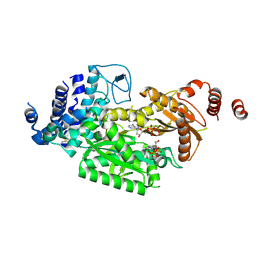 | | fused To-MtbCsm1 with 2dATP | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A),CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), MAGNESIUM ION | | 著者 | Li, T, Huo, Y, Jiang, T. | | 登録日 | 2019-06-24 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Mycobacterium tuberculosis CRISPR/Cas system Csm1 holds clues to the evolutionary relationship between DNA polymerase and cyclase activity.
Int.J.Biol.Macromol., 170, 2020
|
|
7XFR
 
 | |
3LDN
 
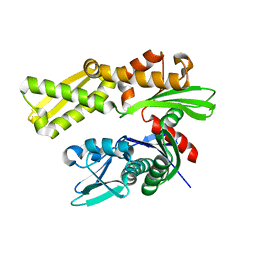 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in apo form | | 分子名称: | 78 kDa glucose-regulated protein | | 著者 | Dokurno, P, Surgenor, A.E, Shaw, T, Macias, A.T, Massey, A.J, Williamson, D.S. | | 登録日 | 2010-01-13 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Adenosine-Derived Inhibitors of 78 kDa Glucose Regulated Protein (Grp78) ATPase: Insights into Isoform Selectivity.
J.Med.Chem., 54, 2011
|
|
3LDQ
 
 | | Crystal structure of HSC70/BAG1 in complex with small molecule inhibitor | | 分子名称: | 8-[(quinolin-2-ylmethyl)amino]adenosine, BAG family molecular chaperone regulator 1, Heat shock cognate 71 kDa protein | | 著者 | Dokurno, P, Surgenor, A.E, Shaw, T, Macias, A.T, Massey, A.J, Williamson, D.S. | | 登録日 | 2010-01-13 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Adenosine-Derived Inhibitors of 78 kDa Glucose Regulated Protein (Grp78) ATPase: Insights into Isoform Selectivity.
J.Med.Chem., 54, 2011
|
|
3LDL
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with ATP | | 分子名称: | 78 kDa glucose-regulated protein, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | 著者 | Dokurno, P, Surgenor, A.E, Shaw, T, Macias, A.T, Massey, A.J, Williamson, D.S. | | 登録日 | 2010-01-13 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Adenosine-Derived Inhibitors of 78 kDa Glucose Regulated Protein (Grp78) ATPase: Insights into Isoform Selectivity.
J.Med.Chem., 54, 2011
|
|
3PMI
 
 | | PWWP Domain of Human Mutated Melanoma-Associated Antigen 1 | | 分子名称: | DI(HYDROXYETHYL)ETHER, PWWP domain-containing protein MUM1, SULFATE ION, ... | | 著者 | Lam, R, Zeng, H, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | 登録日 | 2010-11-17 | | 公開日 | 2010-12-15 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | Structural and histone binding ability characterizations of human PWWP domains.
Plos One, 6, 2011
|
|
3PFS
 
 | | PWWP Domain of Human Bromodomain and PHD finger-containing protein 3 | | 分子名称: | Bromodomain and PHD finger-containing protein 3, SULFATE ION, ZINC ION | | 著者 | Lam, R, Zeng, H, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | 登録日 | 2010-10-29 | | 公開日 | 2010-11-10 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and histone binding ability characterizations of human PWWP domains.
Plos One, 6, 2011
|
|
3Q4U
 
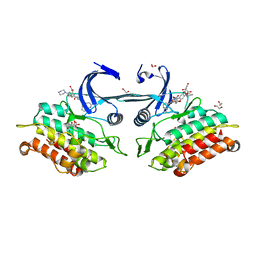 | | Crystal structure of the ACVR1 kinase domain in complex with LDN-193189 | | 分子名称: | 1,2-ETHANEDIOL, 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Activin receptor type-1, ... | | 著者 | Chaikuad, A, Sanvitale, C, Cooper, C.D.O, Mahajan, P, Daga, N, Petrie, K, Alfano, I, Gileadi, O, Fedorov, O, Allerston, C.K, Krojer, T, Vollmar, M, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | 登録日 | 2010-12-24 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | A new class of small molecule inhibitor of BMP signaling.
Plos One, 8, 2013
|
|
7TVA
 
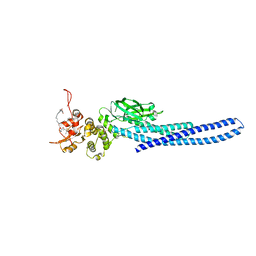 | | Stat5a Core in complex with AK2292 | | 分子名称: | DI(HYDROXYETHYL)ETHER, MALONATE ION, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N-(5-{2-[(3R)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}pent-4-yn-1-yl)-N-methyl-N~3~-[4-(1,3-thiazol-2-yl)phenyl]-beta-alaninamide, ... | | 著者 | Meagher, J.L, Stuckey, J.A. | | 登録日 | 2022-02-04 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.835 Å) | | 主引用文献 | A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo.
Nat.Chem.Biol., 19, 2023
|
|
7TVB
 
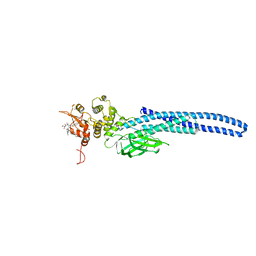 | | Stat5A Core in Complex with AK305 | | 分子名称: | GLYCEROL, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N,N-dimethyl-N~3~-[4-(1,3-thiazol-2-yl)phenyl]-beta-alaninamide, Signal transducer and activator of transcription 5A | | 著者 | Meagher, J.L, Stuckey, J.A. | | 登録日 | 2022-02-04 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.653 Å) | | 主引用文献 | A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo.
Nat.Chem.Biol., 19, 2023
|
|
4G2M
 
 | |
4G0B
 
 | |
4FX4
 
 | |
7C8U
 
 | | The crystal structure of COVID-19 main protease in complex with GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | 著者 | Luan, X, Shang, W, Wang, Y, Yin, W, Jiang, Y, Feng, S, Wang, Y, Liu, M, Zhou, R, Zhang, Z, Wang, F, Cheng, W, Gao, M, Wang, H, Wu, W, Tian, R, Tian, Z, Jin, Y, Jiang, H.W, Zhang, L, Xu, H.E, Zhang, S. | | 登録日 | 2020-06-03 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | The crystal structure of COVID-19 main protease in complex with GC376
To Be Published
|
|
7V9P
 
 | |
7V9Q
 
 | |
7V9O
 
 | |
7V9N
 
 | | Crystal structure of the lanthipeptide zinc-metallopeptidase EryP from saccharopolyspora erythraea in closed state | | 分子名称: | ACETATE ION, Alanine aminopeptidase, CALCIUM ION, ... | | 著者 | Zhao, C, Zhao, N.L, Bao, R. | | 登録日 | 2021-08-26 | | 公開日 | 2022-05-11 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Conformational remodeling enhances activity of lanthipeptide zinc-metallopeptidases.
Nat.Chem.Biol., 18, 2022
|
|
2Z60
 
 | | Crystal Structure of the T315I Mutant of Abl kinase bound with PPY-A | | 分子名称: | 5-[3-(2-METHOXYPHENYL)-1H-PYRROLO[2,3-B]PYRIDIN-5-YL]-N,N-DIMETHYLPYRIDINE-3-CARBOXAMIDE, Proto-oncogene tyrosine-protein kinase ABL1 | | 著者 | Zhou, T, Dalgarno, D, Zhu, X. | | 登録日 | 2007-07-21 | | 公開日 | 2007-09-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structure of the T315I Mutant of Abl Kinase
Chem.Biol.Drug Des., 70, 2007
|
|
4GIW
 
 | |
