2FPK
 
 | | RadA recombinase in complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA repair and recombination protein radA, MAGNESIUM ION, ... | | 著者 | Wu, Y, Qian, X, He, Y, Moya, I.A, Luo, Y. | | 登録日 | 2006-01-16 | | 公開日 | 2006-01-31 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of Methanococcus Voltae Rada in Complex with Adp: hydrolysis-induced conformational change
Biochemistry, 44, 2005
|
|
2FPM
 
 | | RadA recombinase in complex with AMP-PNP and high concentration of K+ | | 分子名称: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Wu, Y, Qian, X, He, Y, Moya, I.A, Luo, Y. | | 登録日 | 2006-01-16 | | 公開日 | 2006-01-31 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Methanoccocus Voltae Rada in Complex with Adp: hydrolysis-induced conformational change
Biochemistry, 44, 2005
|
|
6O9M
 
 | | Structure of the human apo TFIIH | | 分子名称: | CDK-activating kinase assembly factor MAT1, General transcription factor IIH subunit 1, General transcription factor IIH subunit 2, ... | | 著者 | Yan, C.L, Dodd, T, He, Y, Tainer, J.A, Tsutakawa, S.E, Ivanov, I. | | 登録日 | 2019-03-14 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Transcription preinitiation complex structure and dynamics provide insight into genetic diseases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2F1J
 
 | | Recombinase in Complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA repair and recombination protein radA, MAGNESIUM ION | | 著者 | Qian, X, He, Y, Wu, Y, Luo, Y. | | 登録日 | 2005-11-14 | | 公開日 | 2006-05-30 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Asp302 determines potassium dependence of a RadA recombinase from Methanococcus voltae.
J.Mol.Biol., 360, 2006
|
|
2VSN
 
 | | Structure and topological arrangement of an O-GlcNAc transferase homolog: insight into molecular control of intracellular glycosylation | | 分子名称: | URIDINE-5'-DIPHOSPHATE, XCOGT | | 著者 | Martinez-Fleites, C, Macauley, M.S, He, Y, Shen, D, Vocadlo, D, Davies, G.J. | | 登録日 | 2008-04-28 | | 公開日 | 2008-06-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure of an O-Glcnac Transferase Homolog Provides Insight Into Intracellular Glycosylation.
Nat.Struct.Mol.Biol., 15, 2008
|
|
4GJT
 
 | | complex structure of nectin-4 bound to MV-H | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin glycoprotein, Poliovirus receptor-related protein 4 | | 著者 | Zhang, X, Lu, G, Qi, J, Li, Y, He, Y, Xu, X, Shi, J, Zhang, C, Yan, J, Gao, G.F. | | 登録日 | 2012-08-10 | | 公開日 | 2012-10-10 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3.1001 Å) | | 主引用文献 | Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4
Nat.Struct.Mol.Biol., 20, 2013
|
|
2GDJ
 
 | | Delta-62 RADA recombinase in complex with AMP-PNP and magnesium | | 分子名称: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Wu, Y, Qian, X, He, Y, Luo, Y. | | 登録日 | 2006-03-16 | | 公開日 | 2006-05-16 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Rad51/RadA N-Terminal Domain Activates Nucleoprotein Filament ATPase Activity.
Structure, 14, 2006
|
|
2F1I
 
 | | Recombinase in Complex with AMP-PNP | | 分子名称: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Qian, X, He, Y, Wu, Y, Luo, Y. | | 登録日 | 2005-11-14 | | 公開日 | 2006-05-30 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Asp302 determines potassium dependence of a RadA recombinase from Methanococcus voltae.
J.Mol.Biol., 360, 2006
|
|
7TRD
 
 | | Human telomerase catalytic core structure at 3.3 Angstrom | | 分子名称: | Telomerase RNA, partial sequence, Telomerase reverse transcriptase, ... | | 著者 | Liu, B, He, Y, Wang, Y, Song, H, Zhou, Z.H, Feigon, J. | | 登録日 | 2022-01-28 | | 公開日 | 2022-04-20 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of active human telomerase with telomere shelterin protein TPP1.
Nature, 604, 2022
|
|
7TRE
 
 | | Human telomerase catalytic core with shelterin protein TPP1 | | 分子名称: | Adrenocortical dysplasia protein homolog, Telomerase RNA, partial sequence, ... | | 著者 | Liu, B, He, Y, Wang, Y, Song, H, Zhou, Z.H, Feigon, J. | | 登録日 | 2022-01-28 | | 公開日 | 2022-04-20 | | 最終更新日 | 2025-06-04 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of active human telomerase with telomere shelterin protein TPP1.
Nature, 604, 2022
|
|
7TRC
 
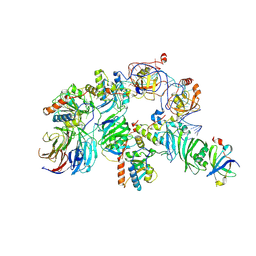 | | Human telomerase H/ACA RNP at 3.3 Angstrom | | 分子名称: | H/ACA ribonucleoprotein complex subunit 1, H/ACA ribonucleoprotein complex subunit 2, H/ACA ribonucleoprotein complex subunit 3, ... | | 著者 | Liu, B, He, Y, Wang, Y, Song, H, Zhou, Z.H, Feigon, J. | | 登録日 | 2022-01-28 | | 公開日 | 2022-04-20 | | 最終更新日 | 2025-06-04 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of active human telomerase with telomere shelterin protein TPP1.
Nature, 604, 2022
|
|
7TRF
 
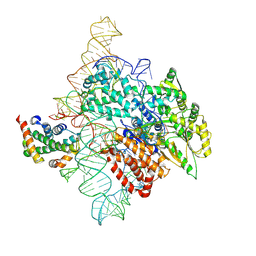 | | Human telomerase catalytic core RNP with H2A/H2B | | 分子名称: | Histone H2A, Histone H2B type 1-C/E/F/G/I, Telomerase RNA, ... | | 著者 | Liu, B, He, Y, Wang, Y, Song, H, Zhou, Z.H, Feigon, J. | | 登録日 | 2022-01-28 | | 公開日 | 2022-04-20 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structure of active human telomerase with telomere shelterin protein TPP1.
Nature, 604, 2022
|
|
2F1H
 
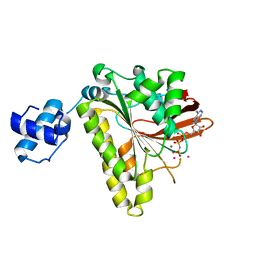 | | RECOMBINASE IN COMPLEX WITH AMP-PNP and Potassium | | 分子名称: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Qian, X, He, Y, Wu, Y, Luo, Y. | | 登録日 | 2005-11-14 | | 公開日 | 2006-05-30 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Asp302 determines potassium dependence of a RadA recombinase from Methanococcus voltae.
J.Mol.Biol., 360, 2006
|
|
5DGY
 
 | | Crystal structure of rhodopsin bound to visual arrestin | | 分子名称: | Endolysin,Rhodopsin,S-arrestin | | 著者 | Zhou, X.E, Gao, X, Kang, Y, He, Y, de Waal, P.W, Suino-Powell, K.M, Wang, M, Melcher, K, Xu, H.E. | | 登録日 | 2015-08-28 | | 公開日 | 2016-03-23 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (7.7 Å) | | 主引用文献 | X-ray laser diffraction for structure determination of the rhodopsin-arrestin complex.
Sci Data, 3, 2016
|
|
5W0P
 
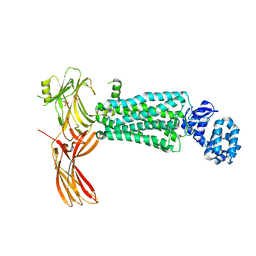 | | Crystal structure of rhodopsin bound to visual arrestin determined by X-ray free electron laser | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endolysin,Rhodopsin,S-arrestin | | 著者 | Zhou, X.E, He, Y, de Waal, P.W, Gao, X, Kang, Y, Van Eps, N, Yin, Y, Pal, K, Goswami, D, White, T.A, Barty, A, Latorraca, N.R, Chapman, H.N, Hubbell, W.L, Dror, R.O, Stevens, R.C, Cherezov, V, Gurevich, V.V, Griffin, P.R, Ernst, O.P, Melcher, K, Xu, H.E. | | 登録日 | 2017-05-31 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3.013 Å) | | 主引用文献 | Identification of Phosphorylation Codes for Arrestin Recruitment by G Protein-Coupled Receptors.
Cell, 170, 2017
|
|
5YB3
 
 | | Crystal structure of HP23L/N36 | | 分子名称: | Envelope glycoprotein, HP23L | | 著者 | Zhang, X, Wang, X, He, Y. | | 登録日 | 2017-09-03 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.043 Å) | | 主引用文献 | Structural Insights into the Mechanisms of Action of Short-Peptide HIV-1 Fusion Inhibitors Targeting the Gp41 Pocket
Front Cell Infect Microbiol, 8, 2018
|
|
4LYC
 
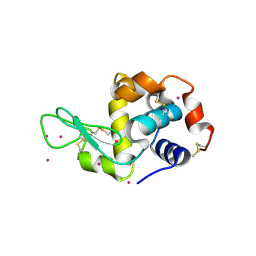 | | Cd ions within a lysoyzme single crystal | | 分子名称: | CADMIUM ION, Lysozyme C | | 著者 | Wei, H, House, S, Wu, J, Zhang, J, Wang, Z, He, Y, Gao, Y.-G, Robinson, H, Li, W, Zuo, J.-M, Robertson, I.M, Lu, Y. | | 登録日 | 2013-07-30 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Enhanced and tunable fluorescent quantum dots within a single crystal of protein
TO BE PUBLISHED
|
|
3O5X
 
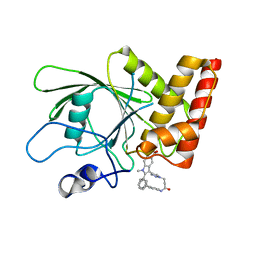 | | Crystal structure of the oncogenic tyrosine phosphatase SHP2 complexed with a salicylic acid-based small molecule inhibitor | | 分子名称: | 3-{1-[3-(biphenyl-4-ylamino)-3-oxopropyl]-1H-1,2,3-triazol-4-yl}-6-hydroxy-1-methyl-2-phenyl-1H-indole-5-carboxylic acid, Tyrosine-protein phosphatase non-receptor type 11 | | 著者 | Zhang, Z.-Y, Zhang, X, He, Y, Liu, S, Yu, Z, Jiang, Z, Yang, Z, Dong, Y, Nabinger, S.C, Wu, L, Gunawan, A.M, Wang, L, Chan, R.J. | | 登録日 | 2010-07-28 | | 公開日 | 2010-08-11 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Salicylic acid based small molecule inhibitor for the oncogenic Src homology-2 domain containing protein tyrosine phosphatase-2 (SHP2).
J.Med.Chem., 53, 2010
|
|
4LYB
 
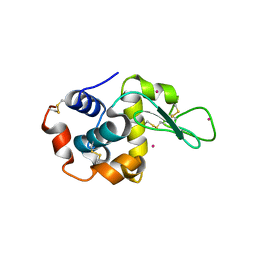 | | CdS within a lysoyzme single crystal | | 分子名称: | CADMIUM ION, Lysozyme C | | 著者 | Wei, H, House, S, Wu, J, Zhang, J, Wang, Z, He, Y, Gao, Y.-G, Robinson, H, Li, W, Zuo, J.-M, Robertson, I.M, Lu, Y. | | 登録日 | 2013-07-30 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Enhanced and tunable fluorescent quantum dots within a single crystal of protein
TO BE PUBLISHED
|
|
5YB2
 
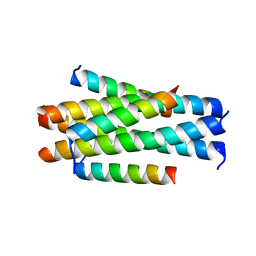 | | Crystal structure of LP-11/N44 | | 分子名称: | Envelope glycoprotein, LP-11 | | 著者 | Zhang, X, Wang, X, He, Y. | | 登録日 | 2017-09-03 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structural Insights into the Mechanisms of Action of Short-Peptide HIV-1 Fusion Inhibitors Targeting the Gp41 Pocket
Front Cell Infect Microbiol, 8, 2018
|
|
5YC0
 
 | | Crystal structure of LP-46/N44 | | 分子名称: | Envelope glycoprotein, LP-46 | | 著者 | Zhang, X, Wang, X, He, Y. | | 登録日 | 2017-09-05 | | 公開日 | 2018-02-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Exceptional potency and structural basis of a T1249-derived lipopeptide fusion inhibitor against HIV-1, HIV-2, and simian immunodeficiency virus
J. Biol. Chem., 293, 2018
|
|
4QKQ
 
 | |
5YB4
 
 | | Crystal structure of HP23LN36KR | | 分子名称: | HP23L, N36KR | | 著者 | Zhang, X, Wang, X, He, Y. | | 登録日 | 2017-09-03 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Insights into the Mechanisms of Action of Short-Peptide HIV-1 Fusion Inhibitors Targeting the Gp41 Pocket
Front Cell Infect Microbiol, 8, 2018
|
|
2I1Q
 
 | | RadA Recombinase in complex with Calcium | | 分子名称: | CALCIUM ION, DNA repair and recombination protein radA, MAGNESIUM ION, ... | | 著者 | Qian, X, He, Y, Ma, X, Fodje, M.N, Grochulski, P, Luo, Y. | | 登録日 | 2006-08-14 | | 公開日 | 2006-10-24 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Calcium Stiffens Archaeal Rad51 Recombinase from Methanococcus voltae for Homologous Recombination.
J.Biol.Chem., 281, 2006
|
|
5W66
 
 | | RNA polymerase I Initial Transcribing Complex State 3 | | 分子名称: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | 著者 | Han, Y, He, Y. | | 登録日 | 2017-06-16 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
