8HVW
 
 | |
6LU7
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor N3 | | 分子名称: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | 著者 | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | 登録日 | 2020-01-26 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
5WUU
 
 | |
7BQY
 
 | | THE CRYSTAL STRUCTURE OF COVID-19 MAIN PROTEASE IN COMPLEX WITH AN INHIBITOR N3 at 1.7 angstrom | | 分子名称: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | 著者 | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | 登録日 | 2020-03-26 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
4MBS
 
 | | Crystal Structure of the CCR5 Chemokine Receptor | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4,4-difluoro-N-[(1S)-3-{(3-exo)-3-[3-methyl-5-(propan-2-yl)-4H-1,2,4-triazol-4-yl]-8-azabicyclo[3.2.1]oct-8-yl}-1-phenylpropyl]cyclohexanecarboxamide, Chimera protein of C-C chemokine receptor type 5 and Rubredoxin, ... | | 著者 | Tan, Q, Zhu, Y, Han, G.W, Li, J, Fenalti, G, Liu, H, Cherezov, V, Stevens, R.C, GPCR Network (GPCR), Zhao, Q, Wu, B. | | 登録日 | 2013-08-19 | | 公開日 | 2013-09-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex.
Science, 341, 2013
|
|
5BOA
 
 | | Crystal Structure of the Meningitis Pathogen Streptococcus suis adhesion Fhb bound to the disaccharide receptor Gb2 | | 分子名称: | Translation initiation factor 2 (IF-2 GTPase), alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose | | 著者 | Zhang, C, Yu, Y, Yang, M, Jiang, Y. | | 登録日 | 2015-05-27 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.708 Å) | | 主引用文献 | Structural basis of the interaction between the meningitis pathogen Streptococcus suis adhesin Fhb and its human receptor.
Febs Lett., 590, 2016
|
|
4G2Y
 
 | | Crystal structure of PDE5A complexed with its inhibitor | | 分子名称: | 2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}-3,5,6,7-tetrahydro-4H-cyclopenta[d]pyrimidin-4-one, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | 著者 | Ren, J, Chen, T.T, Xu, Y.C. | | 登録日 | 2012-07-13 | | 公開日 | 2013-06-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Design, synthesis, and pharmacological evaluation of monocyclic pyrimidinones as novel inhibitors of PDE5.
J.Med.Chem., 55, 2012
|
|
4G2W
 
 | | Crystal structure of PDE5A in complex with its inhibitor | | 分子名称: | 5,6-diethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | 著者 | Ren, J, Chen, T.T, Xu, Y.C. | | 登録日 | 2012-07-13 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Design, synthesis, and pharmacological evaluation of monocyclic pyrimidinones as novel inhibitors of PDE5.
J.Med.Chem., 55, 2012
|
|
6LU5
 
 | | Crystal structure of BPTF-BRD with ligand DCBPin5 bound | | 分子名称: | 6-(1H-indol-5-yl)-N-methyl-2-methylsulfonyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | 著者 | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | 登録日 | 2020-01-25 | | 公開日 | 2021-04-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.86527729 Å) | | 主引用文献 | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
6LU6
 
 | | Crystal structure of BPTF-BRD with ligand DCBPin5-2 bound | | 分子名称: | 6-[1-[3-(dimethylamino)propyl]indol-5-yl]-2-methylsulfonyl-N-propyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | 著者 | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | 登録日 | 2020-01-26 | | 公開日 | 2021-04-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.970063 Å) | | 主引用文献 | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
5EFX
 
 | | Crystal structure of Rho GTPase regulator | | 分子名称: | Rho guanine nucleotide exchange factor 2 | | 著者 | Jiang, Y, Ouyang, S, Liu, Z.J. | | 登録日 | 2015-10-26 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.451 Å) | | 主引用文献 | Crystal structure of hGEF-H1 PH domain provides insight into incapability in phosphoinositide binding
Biochem.Biophys.Res.Commun., 471, 2016
|
|
6LT0
 
 | | cryo-EM structure of C9ORF72-SMCR8-WDR41 | | 分子名称: | Guanine nucleotide exchange C9orf72, Guanine nucleotide exchange protein SMCR8, WD repeat-containing protein 41 | | 著者 | Tang, D, Sheng, J, Xu, L, Zhan, X, Yan, C, Qi, S. | | 登録日 | 2020-01-21 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structure of C9ORF72-SMCR8-WDR41 reveals the role as a GAP for Rab8a and Rab11a.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4IA0
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | 分子名称: | 5-bromo-2-{2-ethoxy-5-[(4-methylpiperazin-1-yl)sulfonyl]phenyl}-6-octylpyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | 著者 | Ren, J, Chen, T, Xu, Y. | | 登録日 | 2012-12-05 | | 公開日 | 2014-01-01 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Exploration of the 5-bromopyrimidin-4(3H)-ones as potent inhibitors of PDE5.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4XZ7
 
 | | Crystal structure of a TGase | | 分子名称: | Putative uncharacterized protein | | 著者 | Yu, J, Ge, J, Yang, M. | | 登録日 | 2015-02-04 | | 公開日 | 2015-06-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | Functional and Structural Characterization of the Antiphagocytic Properties of a Novel Transglutaminase from Streptococcus suis
J.Biol.Chem., 290, 2015
|
|
6GJH
 
 | |
8AXW
 
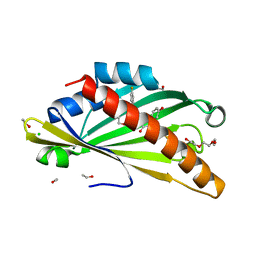 | | The structure of mouse AsterC (GramD1c) with Ezetimibe | | 分子名称: | (3~{R},4~{S})-1-(4-fluorophenyl)-3-[(3~{S})-3-(4-fluorophenyl)-3-oxidanyl-propyl]-4-(4-hydroxyphenyl)azetidin-2-one, CHLORIDE ION, ETHANOL, ... | | 著者 | Fairall, L, Xiao, X, Burger, L, Tontonoz, P, Schwabe, J.W.R. | | 登録日 | 2022-09-01 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Aster-dependent nonvesicular transport facilitates dietary cholesterol uptake.
Science, 382, 2023
|
|
3OZJ
 
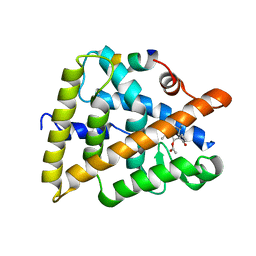 | | Crystal structure of human retinoic X receptor alpha complexed with bigelovin and coactivator SRC-1 | | 分子名称: | (3aR,4S,4aR,7aR,8R,9aS)-4a,8-dimethyl-3-methylidene-2,5-dioxo-2,3,3a,4,4a,5,7a,8,9,9a-decahydroazuleno[6,5-b]furan-4-yl acetate, Retinoic acid receptor RXR-alpha, SRC-1, ... | | 著者 | Zhang, H, Li, L, Chen, L, Hu, L, Shen, X. | | 登録日 | 2010-09-25 | | 公開日 | 2011-02-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure basis of bigelovin as a selective RXR agonist with a distinct binding mode
J.Mol.Biol., 407, 2011
|
|
3PNR
 
 | |
3RIY
 
 | |
3RIG
 
 | |
4EMM
 
 | |
7XZ5
 
 | | GPR119-Gs-LPC complex | | 分子名称: | (4R,7R,18E)-4,7-dihydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphaheptacos-18-en-1-aminium 4-oxide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Xu, P, Huang, S, Jiang, Y, Xu, H.E. | | 登録日 | 2022-06-02 | | 公開日 | 2022-08-24 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural identification of lysophosphatidylcholines as activating ligands for orphan receptor GPR119.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7XZ6
 
 | | GPR119-Gs-APD668 complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Xu, P, Huang, S, Jiang, Y, Xu, H.E. | | 登録日 | 2022-06-02 | | 公開日 | 2022-08-24 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural identification of lysophosphatidylcholines as activating ligands for orphan receptor GPR119.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4EMP
 
 | | Crystal structure of the mutant of ClpP E137A from Staphylococcus aureus | | 分子名称: | ATP-dependent Clp protease proteolytic subunit | | 著者 | Ye, F, Zhang, J, Liu, H, Luo, C, Yang, C.-G. | | 登録日 | 2012-04-12 | | 公開日 | 2013-04-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Helix unfolding/refolding characterizes the functional dynamics of Staphylococcus aureus Clp protease
J.Biol.Chem., 288, 2013
|
|
6EE9
 
 | |
