8F4O
 
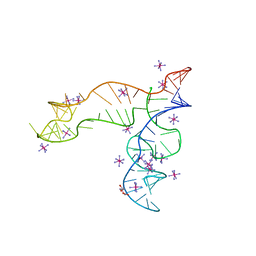 | | Apo structure of the TPP riboswitch aptamer domain | | 分子名称: | IRIDIUM HEXAMMINE ION, TETRAETHYLENE GLYCOL, TPP riboswitch aptamer domain, ... | | 著者 | Lee, H.-K, Wang, Y.-X, Stagno, J.R. | | 登録日 | 2022-11-11 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal structure of Escherichia coli thiamine pyrophosphate-sensing riboswitch in the apo state.
Structure, 31, 2023
|
|
6L0X
 
 | | The First Tudor Domain of PHF20L1 | | 分子名称: | CITRIC ACID, GLYCEROL, PHD finger protein 20-like protein 1 | | 著者 | Lv, M.Q, Gao, J. | | 登録日 | 2019-09-27 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1P
 
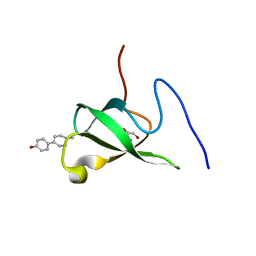 | | Crystal structure of PHF20L1 in complex with Hit 1 | | 分子名称: | 4-(1-methyl-3,6-dihydro-2H-pyridin-4-yl)phenol, GLYCEROL, PHD finger protein 20-like protein 1, ... | | 著者 | Lv, M.Q, Gao, J. | | 登録日 | 2019-09-29 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.231 Å) | | 主引用文献 | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1C
 
 | |
6L1I
 
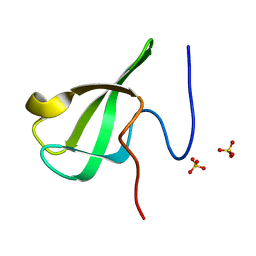 | |
6A60
 
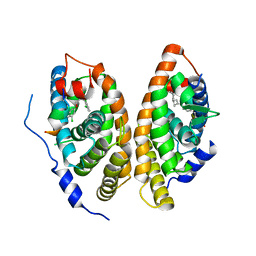 | | Crystal structure of human FXR/RXR-LBD heterodimer bound to GW4064 and 9cRA and SRC1 | | 分子名称: | (9cis)-retinoic acid, 3-[(E)-2-(2-chloro-4-{[3-(2,6-dichlorophenyl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}phenyl)ethenyl]benzoic acid, Bile acid receptor, ... | | 著者 | Wang, N, Liu, J. | | 登録日 | 2018-06-25 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Ligand binding and heterodimerization with retinoid X receptor alpha (RXR alpha ) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding
J. Biol. Chem., 293, 2018
|
|
7N00
 
 | | Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 648-1025 in complex with AUG-alpha | | 分子名称: | ALK and LTK ligand 2, ALK tyrosine kinase receptor | | 著者 | Reshetnyak, A.V, Myasnikov, A.G, Rossi, P, Miller, D.J, Kalodimos, C.G. | | 登録日 | 2021-05-24 | | 公開日 | 2021-11-24 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.27 Å) | | 主引用文献 | Mechanism for the activation of the anaplastic lymphoma kinase receptor.
Nature, 600, 2021
|
|
7MZY
 
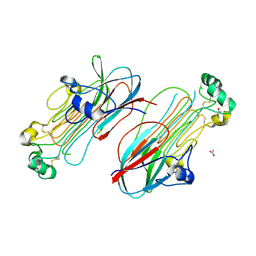 | | Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 673-986 | | 分子名称: | ACETATE ION, ALK tyrosine kinase receptor | | 著者 | Reshetnyak, A.V, Sowaileh, M, Miller, D.J, Rossi, P, Myasnikov, A.G, Kalodimos, C.G. | | 登録日 | 2021-05-24 | | 公開日 | 2021-11-24 | | 最終更新日 | 2021-12-15 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Mechanism for the activation of the anaplastic lymphoma kinase receptor.
Nature, 600, 2021
|
|
7MZZ
 
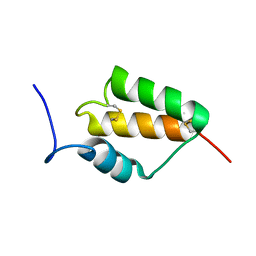 | |
6LAD
 
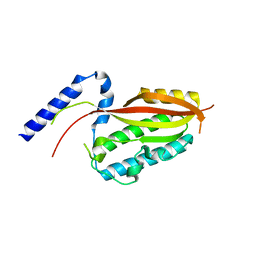 | |
6AGF
 
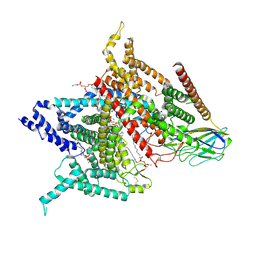 | | Structure of the human voltage-gated sodium channel Nav1.4 in complex with beta1 | | 分子名称: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Pan, X.J, li, Z.Q, Zhou, Q, Shen, H.Z, Wu, K, Huang, X.S, Chen, J.F, Zhang, J.R, Zhu, X.C, Lei, J.L, Xiong, W, Gong, H.P, Xiao, B.L, Yan, N. | | 登録日 | 2018-08-11 | | 公開日 | 2018-10-10 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of the human voltage-gated sodium channel Nav1.4 in complex with beta 1.
Science, 362, 2018
|
|
7MZW
 
 | |
7Z8Z
 
 | |
7YMD
 
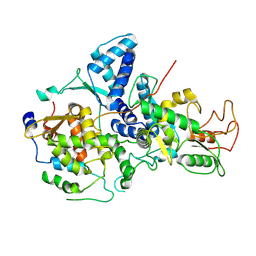 | | Cryo-EM structure of Nse1/3/4 | | 分子名称: | Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosome element 4, Non-structural maintenance of chromosomes element 1 | | 著者 | Qian, L, Jun, Z, Zhenguo, C, Wang, L. | | 登録日 | 2022-07-28 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4.176 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
6L10
 
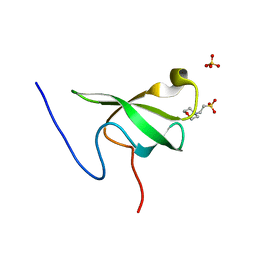 | | PHF20L1 Tudor1 - MES | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PHD finger protein 20-like protein 1, SULFATE ION | | 著者 | Lv, M.Q, Gao, J. | | 登録日 | 2019-09-27 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
1FAD
 
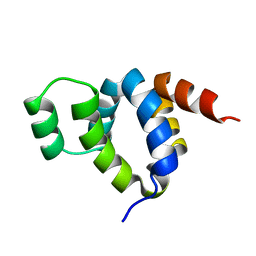 | | DEATH DOMAIN OF FAS-ASSOCIATED DEATH DOMAIN PROTEIN, RESIDUES 89-183 | | 分子名称: | PROTEIN (FADD PROTEIN) | | 著者 | Jeong, E.-J, Bang, S, Lee, T.H, Park, Y.-I, Sim, W.-S, Kim, K.-S. | | 登録日 | 1999-03-23 | | 公開日 | 1999-07-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of FADD death domain. Structural basis of death domain interactions of Fas and FADD.
J.Biol.Chem., 274, 1999
|
|
6VCG
 
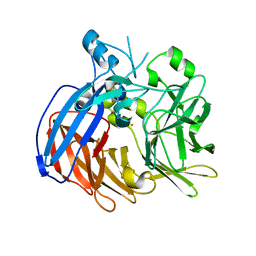 | | Crystal structure of Nitrosotalea devanaterra carotenoid cleavage dioxygenase, cobalt form | | 分子名称: | CHLORIDE ION, COBALT (II) ION, SODIUM ION, ... | | 著者 | Daruwalla, A, Shi, W, Kiser, P.D. | | 登録日 | 2019-12-20 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6L1F
 
 | |
7MZX
 
 | |
6LAF
 
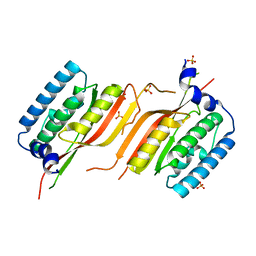 | | Crystal structure of the core domain of Amuc_1100 from Akkermansia muciniphila | | 分子名称: | Amuc_1100, SULFATE ION | | 著者 | Wang, J, Xiang, R, Zhang, M, Wang, M. | | 登録日 | 2019-11-12 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.001 Å) | | 主引用文献 | The variable oligomeric state of Amuc_1100 from Akkermansia muciniphila.
J.Struct.Biol., 212, 2020
|
|
7T0P
 
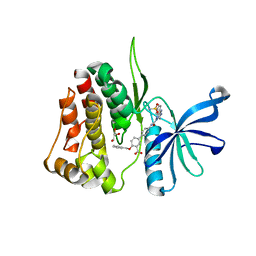 | | JAK2 JH2 IN COMPLEX WITH JAK315 | | 分子名称: | 4'-{[5-amino-3-(4-sulfamoylanilino)-1H-1,2,4-triazole-1-carbonyl]amino}-4-(benzyloxy)[1,1'-biphenyl]-3-carboxylic acid, GLYCEROL, Tyrosine-protein kinase JAK2 | | 著者 | Ippolito, J.A, Liosi, M.-E, Krimmer, S.G, Schlessinger, J, Jorgensen, W.L. | | 登録日 | 2021-11-30 | | 公開日 | 2022-06-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Insights on JAK2 Modulation by Potent, Selective, and Cell-Permeable Pseudokinase-Domain Ligands.
J.Med.Chem., 65, 2022
|
|
7SSB
 
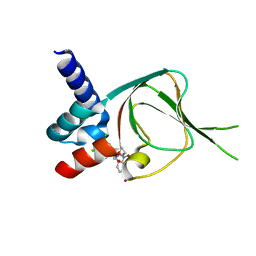 | | Co-structure of PKG1 regulatory domain with compound 33 | | 分子名称: | 4-({(2S,3S)-3-[(1S)-1-(3,5-dichlorophenyl)-2-hydroxyethoxy]-2-phenylpiperidin-1-yl}methyl)-3-nitrobenzoic acid, cGMP-dependent protein kinase 1 | | 著者 | Fischmann, T.O. | | 登録日 | 2021-11-10 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Optimization and Mechanistic Investigations of Novel Allosteric Activators of PKG1 alpha.
J.Med.Chem., 65, 2022
|
|
4R7H
 
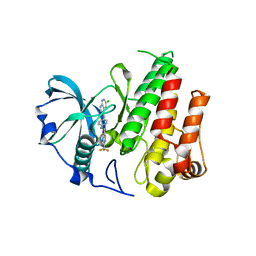 | | Crystal structure of FMS KINASE domain with a small molecular inhibitor, PLX3397 | | 分子名称: | 5-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)methyl]-N-{[6-(trifluoromethyl)pyridin-3-yl]methyl}pyridin-2-amine, Macrophage colony-stimulating factor 1 receptor | | 著者 | Zhang, Y, Zhang, K, Zhang, C. | | 登録日 | 2014-08-27 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8001 Å) | | 主引用文献 | Structure-Guided Blockade of CSF1R Kinase in Tenosynovial Giant-Cell Tumor.
N Engl J Med, 373, 2015
|
|
5DD5
 
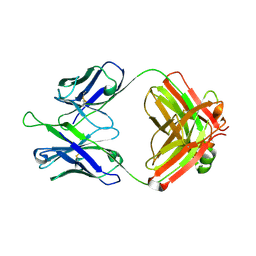 | |
4EJS
 
 | | Structure of yeast elongator subcomplex Elp456 | | 分子名称: | Elongator complex protein 4, Elongator complex protein 5, Elongator complex protein 6 | | 著者 | Lin, Z, Zhao, W, Long, J, Shen, Y. | | 登録日 | 2012-04-07 | | 公開日 | 2012-05-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.606 Å) | | 主引用文献 | Crystal structure of elongator subcomplex Elp4-6
J.Biol.Chem., 287, 2012
|
|
