8WML
 
 | |
7DD8
 
 | |
7DK5
 
 | |
7DK6
 
 | |
7DCC
 
 | |
8WMC
 
 | |
6JE8
 
 | | crystal structure of a beta-N-acetylhexosaminidase | | 分子名称: | Beta-N-acetylhexosaminidase, FORMIC ACID, GLYCEROL, ... | | 著者 | Chen, X, Wang, J.C, Liu, M.J, Yang, W.Y, Wang, Y.Z, Tang, R.P, Zhang, M. | | 登録日 | 2019-02-04 | | 公開日 | 2019-03-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.798 Å) | | 主引用文献 | Crystallographic evidence for substrate-assisted catalysis of beta-N-acetylhexosaminidas from Akkermansia muciniphila.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
5TKK
 
 | |
7NUE
 
 | | Crystal structure of mouse PRMT6 in complex with inhibitor EML736 | | 分子名称: | Protein arginine N-methyltransferase 6, methyl 6-[5-[[~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]carbamimidoyl]amino]pentylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | 著者 | Bonnefond, L, Cavarelli, J. | | 登録日 | 2021-03-11 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7NUD
 
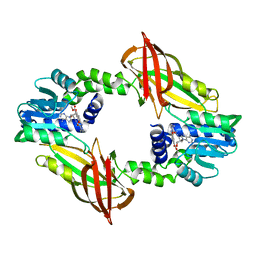 | | Crystal structure of mouse PRMT6 in complex with inhibitor EML734 | | 分子名称: | Protein arginine N-methyltransferase 6, methyl 6-[3-[[~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]carbamimidoyl]amino]propylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | 著者 | Bonnefond, L, Cavarelli, J. | | 登録日 | 2021-03-11 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
5XLT
 
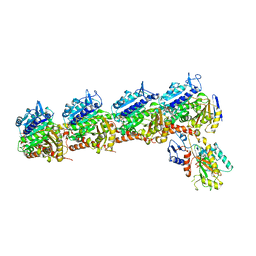 | | The crystal structure of tubulin in complex with 4'-demethylepipodophyllotoxin | | 分子名称: | (5S,5aR,8aR,9R)-9-(3,5-dimethoxy-4-oxidanyl-phenyl)-5-oxidanyl-5a,6,8a,9-tetrahydro-5H-[2]benzofuro[6,5-f][1,3]benzodioxol-8-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Yu, Y, Chen, Q. | | 登録日 | 2017-05-11 | | 公開日 | 2017-09-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.813 Å) | | 主引用文献 | Structure of 4'-demethylepipodophyllotoxin in complex with tubulin provides a rationale for drug design
Biochem. Biophys. Res. Commun., 493, 2017
|
|
7DD2
 
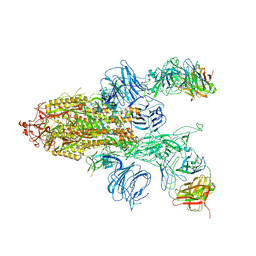 | |
7DK4
 
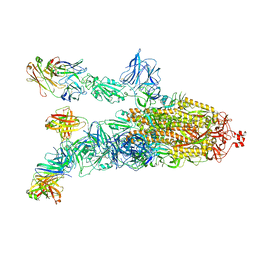 | |
7DDN
 
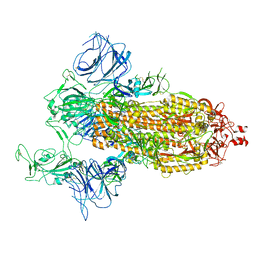 | | SARS-Cov2 S protein at open state | | 分子名称: | Spike glycoprotein | | 著者 | Cong, Y, Liu, C.X. | | 登録日 | 2020-10-29 | | 公開日 | 2020-11-25 | | 最終更新日 | 2021-01-27 | | 実験手法 | ELECTRON MICROSCOPY (6.3 Å) | | 主引用文献 | Development and structural basis of a two-MAb cocktail for treating SARS-CoV-2 infections.
Nat Commun, 12, 2021
|
|
6JEB
 
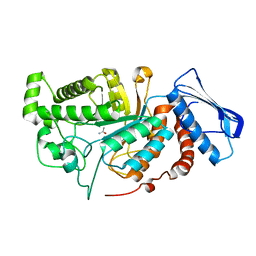 | | crystal structure of a beta-N-acetylhexosaminidase | | 分子名称: | ACETAMIDE, Beta-N-acetylhexosaminidase, ZINC ION | | 著者 | Chen, X, Wang, J.C, Liu, M.J, Yang, W.Y, Wang, Y.Z, Tang, R.P, Zhang, M. | | 登録日 | 2019-02-05 | | 公開日 | 2019-03-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.498 Å) | | 主引用文献 | Crystallographic evidence for substrate-assisted catalysis of beta-N-acetylhexosaminidas from Akkermansia muciniphila.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
7DDD
 
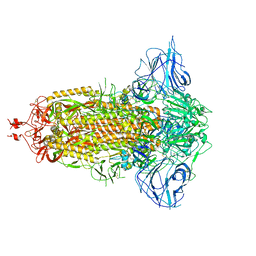 | | SARS-Cov2 S protein at close state | | 分子名称: | Spike glycoprotein | | 著者 | Cong, Y, Liu, C.X. | | 登録日 | 2020-10-28 | | 公開日 | 2020-11-25 | | 最終更新日 | 2021-01-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Development and structural basis of a two-MAb cocktail for treating SARS-CoV-2 infections.
Nat Commun, 12, 2021
|
|
7DK7
 
 | |
7DCX
 
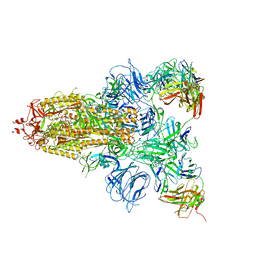 | |
3GBW
 
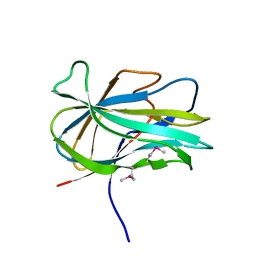 | | Crystal structure of the first PHR domain of the Mouse Myc-binding protein 2 (MYCBP-2) | | 分子名称: | E3 ubiquitin-protein ligase MYCBP2 | | 著者 | Sampathkumar, P, Ozyurt, S.A, Wasserman, S.R, Klemke, R.L, Miller, S.A, Bain, K.T, Rutter, M.E, Tarun, G, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-02-20 | | 公開日 | 2009-03-24 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Structures of PHR domains from Mus musculus Phr1 (Mycbp2) explain the loss-of-function mutation (Gly1092-->Glu) of the C. elegans ortholog RPM-1.
J.Mol.Biol., 397, 2010
|
|
7X2H
 
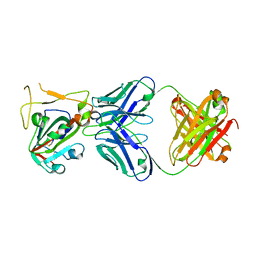 | |
5TKJ
 
 | | Structure of vaccine-elicited diverse HIV-1 neutralizing antibody vFP1.01 in complex with HIV-1 fusion peptide residue 512-519 | | 分子名称: | HIV-1 fusion peptide residue 512-519, SULFATE ION, vFP1.01 chimeric mouse antibody heavy chain, ... | | 著者 | Xu, K, Liu, K, Kwong, P.D. | | 登録日 | 2016-10-06 | | 公開日 | 2018-04-04 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.118 Å) | | 主引用文献 | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
4NOO
 
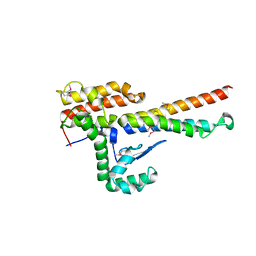 | | Molecular mechanism for self-protection against type VI secretion system in Vibrio cholerae | | 分子名称: | Putative uncharacterized protein, VgrG protein | | 著者 | Yang, X, Xu, M, Jiang, T, Fan, Z. | | 登録日 | 2013-11-20 | | 公開日 | 2014-04-09 | | 最終更新日 | 2014-04-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Molecular mechanism for self-protection against the type VI secretion system in Vibrio cholerae.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4YZW
 
 | | Crystal structure of AgPPO8 | | 分子名称: | AGAP004976-PA, COPPER (II) ION | | 著者 | Hu, Y. | | 登録日 | 2015-03-25 | | 公開日 | 2015-12-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The structure of a prophenoloxidase (PPO) from Anopheles gambiae provides new insights into the mechanism of PPO activation.
Bmc Biol., 14, 2016
|
|
7YL7
 
 | | Structure of hIAPP-TF-type3 | | 分子名称: | Islet amyloid polypeptide | | 著者 | Li, D, Zhang, X. | | 登録日 | 2022-07-25 | | 公開日 | 2022-12-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | A new polymorphism of human amylin fibrils with similar protofilaments and a conserved core.
Iscience, 25, 2022
|
|
5EP6
 
 | | The crystal structure of NAP1 in complex with TBK1 | | 分子名称: | 5-azacytidine-induced protein 2, GLYCEROL, Serine/threonine-protein kinase TBK1 | | 著者 | Li, F, Xie, X, Liu, J, Pan, L. | | 登録日 | 2015-11-11 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.451 Å) | | 主引用文献 | Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins.
Nat Commun, 7, 2016
|
|
