3GZH
 
 | |
6UUF
 
 | | Crystal structure of a Nudix Hydrolase from M. Smegmatis, RenU | | 分子名称: | Nudix Hydrolase, RenU | | 著者 | Wright, K.M, Yoder, J, Shoemaker, S, Hernandez, A, Iheanacho, A, Marques, I, Amzel, M.L, Gabelli, S.B. | | 登録日 | 2019-10-30 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of RenU
To Be Published
|
|
4JLF
 
 | | Inhibitor resistant (R220A) substitution in the Mycobacterium tuberculosis beta-lactamase | | 分子名称: | Beta-lactamase, PHOSPHATE ION | | 著者 | Hazra, S, Kurz, S, Blanchard, J, Bonomo, R. | | 登録日 | 2013-03-12 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Can inhibitor-resistant substitutions in the Mycobacterium tuberculosis beta-Lactamase BlaC lead to clavulanate resistance?: a biochemical rationale for the use of beta-lactam-beta-lactamase inhibitor combinations.
Antimicrob.Agents Chemother., 57, 2013
|
|
4GZR
 
 | | Crystal structure of the Mycobacterium tuberculosis H37Rv EsxOP (Rv2346c-Rv2347c) complex in space group C2221 | | 分子名称: | ESAT-6-like protein 6, ESAT-6-like protein 7, SULFATE ION | | 著者 | Arbing, M.A, Chan, S, He, Q, Harris, L, Zhou, T.T, Kuo, E, Ahn, C.J, Eisenberg, D, TB Structural Genomics Consortium (TBSGC), Integrated Center for Structure and Function Innovation (ISFI) | | 登録日 | 2012-09-06 | | 公開日 | 2012-09-26 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.553 Å) | | 主引用文献 | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
4I0X
 
 | | Crystal structure of the Mycobacterum abscessus EsxEF (Mab_3112-Mab_3113) complex | | 分子名称: | BETA-MERCAPTOETHANOL, ESAT-6-like protein MAB_3112, ESAT-6-like protein MAB_3113, ... | | 著者 | Arbing, M.A, Chan, S, Lu, J, Kuo, E, Harris, L, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2012-11-19 | | 公開日 | 2013-01-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.956 Å) | | 主引用文献 | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
4RAK
 
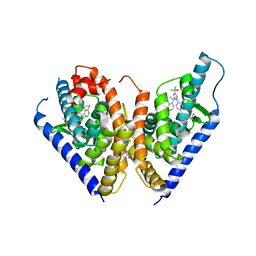 | |
3S0M
 
 | | A Structural Element that Modulates Proton-Coupled Electron Transfer in Oxalate Decarboxylase | | 分子名称: | 1,2-ETHANEDIOL, CARBONATE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Saylor, B.T, Reinhardt, L.A, Lu, Z, Shukla, M.S, Cleland, W.W, Allen, K.N, Richards, N.G.J. | | 登録日 | 2011-05-13 | | 公開日 | 2012-04-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | A structural element that facilitates proton-coupled electron transfer in oxalate decarboxylase.
Biochemistry, 51, 2012
|
|
4QHC
 
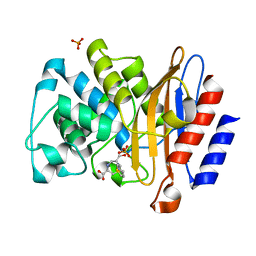 | | Structure of M.Tuberculosis Betalactamase (Blac) with inhibitor having novel mechanism | | 分子名称: | (3R,6R,7S)-7-[(2R,3aR)-hexahydropyrazolo[1,5-c][1,3]thiazin-2-yl]-6-(hydroxymethyl)-1,4-thiazepane-3-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | 著者 | Hazra, S, Blanchard, J. | | 登録日 | 2014-05-28 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.899 Å) | | 主引用文献 | Kinetic and Structural Characterization of the Interaction of 6-Methylidene Penem 2 with the beta-Lactamase from Mycobacterium tuberculosis.
Biochemistry, 54, 2015
|
|
4I1F
 
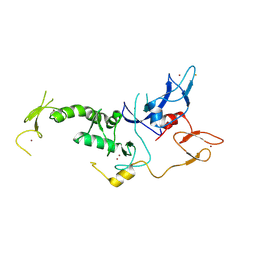 | |
4I1H
 
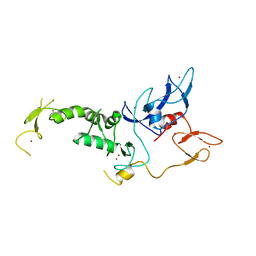 | | Structure of Parkin E3 ligase | | 分子名称: | E3 ubiquitin-protein ligase parkin, ZINC ION | | 著者 | Lougheed, J.C, Brecht, E, Yao, N.H. | | 登録日 | 2012-11-20 | | 公開日 | 2013-06-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases.
Nat Commun, 4, 2013
|
|
3KUT
 
 | |
3KUS
 
 | |
3KUR
 
 | |
6D7G
 
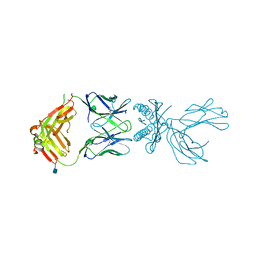 | | Structure of 5F3 TCR in complex with HLA-A2/MART-1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MART1 PEPTIDE-BETA-2-MICROGLOBULIN-HLA-A*02 CHIMERA, T-CELL RECEPTOR GAMMA VARIABLE 8,T-CELL RECEPTOR GAMMA-2 CHAIN C REGION, ... | | 著者 | Roy, S, Adams, E.J. | | 登録日 | 2018-04-24 | | 公開日 | 2019-01-23 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Generation and molecular recognition of melanoma-associated antigen-specific human gamma delta T cells.
Sci Immunol, 3, 2018
|
|
8GAT
 
 | | Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v), based on consensus cryo-EM map with only Fab 1G01 resolved | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 1G01, heavy chain, ... | | 著者 | Tsybovsky, Y, Lederhofer, J, Kwong, P.D, Kanekiyo, M. | | 登録日 | 2023-02-23 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Protective human monoclonal antibodies target conserved sites of vulnerability on the underside of influenza virus neuraminidase.
Immunity, 57, 2024
|
|
8GAU
 
 | | Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 1G01, heavy chain, ... | | 著者 | Tsybovsky, Y, Lederhofer, J, Kwong, P.D, Kanekiyo, M. | | 登録日 | 2023-02-23 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Protective human monoclonal antibodies target conserved sites of vulnerability on the underside of influenza virus neuraminidase.
Immunity, 57, 2024
|
|
8GAV
 
 | | Structure of human NDS.3 Fab in complex with influenza virus neuraminidase from A/Darwin/09/2021 (H3N2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab NDS.3, heavy chain, ... | | 著者 | Tsybovsky, Y, Lederhofer, J, Kwong, P.D, Kanekiyo, M. | | 登録日 | 2023-02-23 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Protective human monoclonal antibodies target conserved sites of vulnerability on the underside of influenza virus neuraminidase.
Immunity, 57, 2024
|
|
2WQA
 
 | | Complex of TTR and RBP4 and Oleic Acid | | 分子名称: | OLEIC ACID, RETINOL-BINDING PROTEIN 4, SULFATE ION, ... | | 著者 | Nanao, M, Mercer, D, Nguyen, L, Buckley, D, Stout, T.J. | | 登録日 | 2009-08-14 | | 公開日 | 2010-09-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Crystal Structure of Rbp4 Bound to Linoleic Acid and Ttr
To be Published
|
|
7GG6
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-477dc5b7-2 (Mpro-x12171) | | 分子名称: | (4R)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.599 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GG7
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with CHO-MSK-00c5269a-2 (Mpro-x12177) | | 分子名称: | 2-(1H-benzotriazol-1-yl)-N-[(3-chlorophenyl)methyl]-N-(4-methoxyphenyl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GGM
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-afd4d4fd-2 (Mpro-x12677) | | 分子名称: | 2-(6-chloropyridin-2-yl)-N-(isoquinolin-4-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.839 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GGO
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-8b8a49e1-4 (Mpro-x12682) | | 分子名称: | (4R)-6-chloro-N-[(4R)-2-oxopiperidin-4-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.687 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GGP
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with VLA-UCB-29506327-1 (Mpro-x12686) | | 分子名称: | (1'M,4S)-6-chloro-1'-(isoquinolin-4-yl)-2,3-dihydrospiro[[1]benzopyran-4,4'-imidazolidine]-2',5'-dione, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.902 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GH2
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-090737b9-1 (Mpro-x12735) | | 分子名称: | (4R)-6-chloro-N-(isoquinolin-4-yl)-N-propanoyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GH4
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-00c1612e-1 (Mpro-x12777) | | 分子名称: | 2-(3-chlorophenyl)-N-(6-methoxyisoquinolin-4-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
