7DRP
 
 | | Structure of ATP-grasp ligase PsnB complexed with phosphomimetic variant of minimal precursor, Mg, and ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-grasp domain-containing protein, MAGNESIUM ION, ... | | 著者 | Song, I, Yu, J, Song, W, Kim, S. | | 登録日 | 2020-12-29 | | 公開日 | 2021-09-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Molecular mechanism underlying substrate recognition of the peptide macrocyclase PsnB.
Nat.Chem.Biol., 17, 2021
|
|
7DRM
 
 | | Structure of ATP-grasp ligase PsnB complexed with minimal precursor, Mg, and ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-grasp domain-containing protein, MAGNESIUM ION, ... | | 著者 | Song, I, Yu, J, Song, W, Kim, S. | | 登録日 | 2020-12-29 | | 公開日 | 2021-09-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.28 Å) | | 主引用文献 | Molecular mechanism underlying substrate recognition of the peptide macrocyclase PsnB.
Nat.Chem.Biol., 17, 2021
|
|
7DRN
 
 | | Structure of ATP-grasp ligase PsnB complexed with precursor peptide PsnA2 and AMPPNP | | 分子名称: | ATP-grasp domain-containing protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PsnA214-38, ... | | 著者 | Song, I, Yu, J, Song, W, Kim, S. | | 登録日 | 2020-12-29 | | 公開日 | 2021-09-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.56 Å) | | 主引用文献 | Molecular mechanism underlying substrate recognition of the peptide macrocyclase PsnB.
Nat.Chem.Biol., 17, 2021
|
|
7DRO
 
 | | Structure of ATP-grasp ligase PsnB complexed with minimal precursor | | 分子名称: | ATP-grasp domain-containing protein, PsnA214-38, Precursor peptide | | 著者 | Song, I, Yu, J, Song, W, Kim, S. | | 登録日 | 2020-12-29 | | 公開日 | 2021-09-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Molecular mechanism underlying substrate recognition of the peptide macrocyclase PsnB.
Nat.Chem.Biol., 17, 2021
|
|
4AC1
 
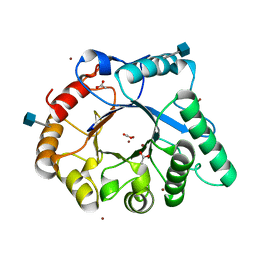 | | The structure of a fungal endo-beta-N-acetylglucosaminidase from glycosyl hydrolase family 18, at 1.3A resolution | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ENDO-N-ACETYL-BETA-D-GLUCOSAMINIDASE, ... | | 著者 | Stals, I, Karkehabadi, S, Devreese, B, Kim, S, Ward, M, Sandgren, M. | | 登録日 | 2011-12-12 | | 公開日 | 2012-08-22 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | High Resolution Crystal Structure of the Endo-N-Acetyl-Beta- D-Glucosaminidase Responsible for the Deglycosylation of Hypocrea Jecorina Cellulases.
Plos One, 7, 2012
|
|
7U5V
 
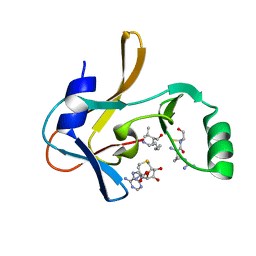 | | Crystal structure of the Mixed Lineage Leukaemia (MLL1) SET Domain with the cofactor product S-Adenosylhomocysteine and Borealin peptide | | 分子名称: | Borealin, Histone-lysine N-methyltransferase 2A, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | An, S, Cho, U.S, Oh, H, Sha, L, Xu, J, Kim, S, Yang, W, An, W, Dou, Y. | | 登録日 | 2022-03-02 | | 公開日 | 2023-09-27 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Non-canonical MLL1 activity regulates centromeric phase separation and genome stability.
Nat.Cell Biol., 25, 2023
|
|
4N8C
 
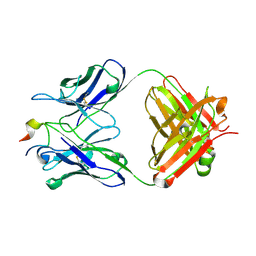 | | Three-dimensional structure of the extracellular domain of Matrix protein 2 of influenza A virus | | 分子名称: | Extracellular domain of influenza Matrix protein 2, Heavy chain of monoclonal antibody, Light chain of monoclonal antibody | | 著者 | Cho, K.J, Seok, J.H, Kim, S, Roose, K, Schepens, B, Fiers, W, Saelens, X, Kim, K.H. | | 登録日 | 2013-10-17 | | 公開日 | 2014-10-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of the extracellular domain of matrix protein 2 of influenza A virus in complex with a protective monoclonal antibody
J.Virol., 89, 2015
|
|
3HQD
 
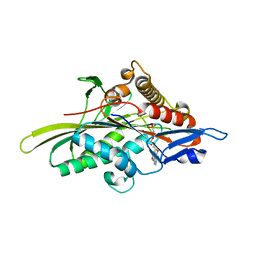 | | Human kinesin Eg5 motor domain in complex with AMPPNP and Mg2+ | | 分子名称: | Kinesin-like protein KIF11, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Parke, C.L, Wojcik, E.J, Kim, S, Worthylake, D.K. | | 登録日 | 2009-06-05 | | 公開日 | 2009-12-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | ATP Hydrolysis in Eg5 Kinesin Involves a Catalytic Two-water Mechanism.
J.Biol.Chem., 285, 2010
|
|
2O0O
 
 | | Crystal structure of TL1A | | 分子名称: | MAGNESIUM ION, TNF superfamily ligand TL1A | | 著者 | Jin, T.C, Kim, S, Guo, F, Howard, A.J, Zhang, Y.Z. | | 登録日 | 2006-11-27 | | 公開日 | 2007-10-30 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | X-ray crystal structure of TNF ligand family member TL1A at 2.1A.
Biochem.Biophys.Res.Commun., 364, 2007
|
|
2P0M
 
 | | Revised structure of rabbit reticulocyte 15S-lipoxygenase | | 分子名称: | (2E)-3-(2-OCT-1-YN-1-YLPHENYL)ACRYLIC ACID, Arachidonate 15-lipoxygenase, FE (II) ION | | 著者 | Choi, J, Chon, J.K, Kim, S, Shin, W. | | 登録日 | 2007-02-28 | | 公開日 | 2007-10-09 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Conformational flexibility in mammalian 15S-lipoxygenase: Reinterpretation of the crystallographic data.
Proteins, 70, 2008
|
|
3GWJ
 
 | | Crystal structure of Antheraea pernyi arylphorin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylphorin, FORMIC ACID, ... | | 著者 | Ryu, K.S, Lee, J.O, Kwon, T.H, Kim, S. | | 登録日 | 2009-04-01 | | 公開日 | 2009-05-05 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | The presence of monoglucosylated N196-glycan is important for the structural stability of storage protein, arylphorin
Biochem.J., 421, 2009
|
|
1TVI
 
 | | Solution structure of TM1509 from Thermotoga maritima: VT1, a NESGC target protein | | 分子名称: | Hypothetical UPF0054 protein TM1509 | | 著者 | Penhoat, C.H, Atreya, H.S, Kim, S, Li, Z, Yee, A, Xiao, R, Murray, D, Arrowsmith, C.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2004-06-29 | | 公開日 | 2005-01-04 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of Thermotoga maritima protein TM1509 reveals a Zn-metalloprotease-like tertiary structure.
J.STRUCT.FUNCT.GENOM., 6, 2005
|
|
1MP6
 
 | |
1P68
 
 | | Solution structure of S-824, a de novo designed four helix bundle | | 分子名称: | De novo designed protein S-824 | | 著者 | Wei, Y, Kim, S, Fela, D, Baum, J, Hecht, M.H. | | 登録日 | 2003-04-29 | | 公開日 | 2003-11-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a de novo protein from a designed combinatorial library.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
1PUL
 
 | | Solution structure for the 21KDa caenorhabditis elegans protein CE32E8.3. NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET WR33 | | 分子名称: | Hypothetical protein C32E8.3 in chromosome I | | 著者 | Tejero, R, Aramini, J.M, Swapna, G.V.T, Monleon, D, Chiang, Y, Macapagal, D, Gunsalus, K.C, Kim, S, Szyperski, T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2003-06-25 | | 公開日 | 2005-06-21 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Backbone 1H, 15N and 13C assignments for the 21 kDa Caenorhabditis elegans homologue of "brain-specific" protein.
J.Biomol.Nmr, 28, 2004
|
|
3VLE
 
 | | Crystal structure of yeast proteasome interacting protein | | 分子名称: | DNA mismatch repair protein HSM3 | | 著者 | Takagi, K, Kim, S, Kato, K, Tanaka, K, Saeki, Y, Mizushima, T. | | 登録日 | 2011-12-01 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structural basis for specific recognition of Rpt1, an ATPase subunit of the 26S proteasome, by a proteasome-dedicated chaperone Hsm3
J.Biol.Chem., 287, 2012
|
|
3VLD
 
 | | Crystal structure of yeast proteasome interacting protein | | 分子名称: | DNA mismatch repair protein HSM3 | | 著者 | Takagi, K, Kim, S, Kato, K, Tanaka, K, Saeki, Y, Mizushima, T. | | 登録日 | 2011-12-01 | | 公開日 | 2012-02-22 | | 最終更新日 | 2012-04-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural basis for specific recognition of Rpt1, an ATPase subunit of the 26S proteasome, by a proteasome-dedicated chaperone Hsm3
J.Biol.Chem., 287, 2012
|
|
3VLF
 
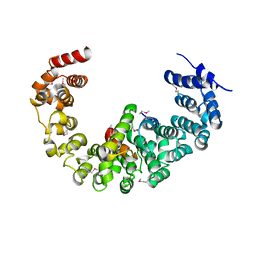 | | Crystal structure of yeast proteasome interacting protein | | 分子名称: | 26S protease regulatory subunit 7 homolog, DNA mismatch repair protein HSM3 | | 著者 | Takagi, K, Kim, S, Kato, K, Tanaka, K, Saeki, Y, Mizushima, T. | | 登録日 | 2011-12-01 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structural basis for specific recognition of Rpt1, an ATPase subunit of the 26S proteasome, by a proteasome-dedicated chaperone Hsm3
J.Biol.Chem., 287, 2012
|
|
1RKJ
 
 | | Solution structure of the complex formed by the two N-terminal RNA-binding domains of nucleolin and a pre-rRNA target | | 分子名称: | 5'-R(*GP*GP*AP*UP*GP*CP*CP*UP*CP*CP*CP*GP*AP*GP*UP*GP*CP*AP*UP*CP*C)-3', Nucleolin | | 著者 | Johansson, C, Finger, L.D, Trantirek, L, Mueller, T.D, Kim, S, Laird-Offringa, I.A, Feigon, J. | | 登録日 | 2003-11-21 | | 公開日 | 2004-04-27 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the complex formed by the two N-terminal RNA-binding domains of nucleolin and a pre-rRNA target.
J.Mol.Biol., 337, 2004
|
|
6JJL
 
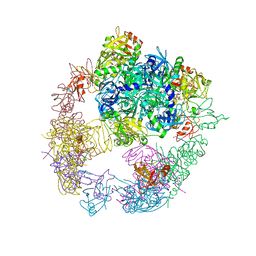 | | Crystal structure of the DegP dodecamer with a modulator | | 分子名称: | CYS-TYR-ARG-LYS-LEU, Periplasmic serine endoprotease DegP | | 著者 | Cho, H, Choi, Y, Lee, H.H, Kim, S. | | 登録日 | 2019-02-26 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (4.2 Å) | | 主引用文献 | Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy.
Commun Biol, 3, 2020
|
|
6JJK
 
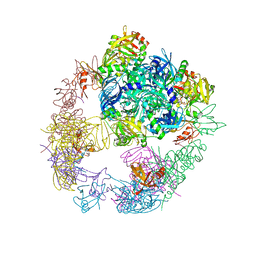 | | Crystal structure of the DegP dodecamer with a modulator | | 分子名称: | CYS-TYR-TYR-LYS-ILE, Periplasmic serine endoprotease DegP | | 著者 | Cho, H, Choi, Y, Lee, H.H, Kim, S. | | 登録日 | 2019-02-26 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy
Commun Biol, 3, 2020
|
|
6JJO
 
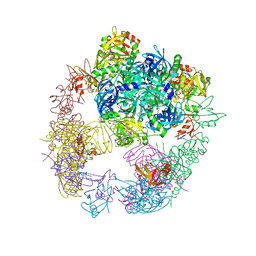 | | Crystal structure of the DegP dodecamer with a modulator | | 分子名称: | Periplasmic serine endoprotease DegP, TMB-CYRKL modulator | | 著者 | Cho, H, Choi, Y, Lee, H.H, Kim, S. | | 登録日 | 2019-02-26 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (4.157 Å) | | 主引用文献 | Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy
Commun Biol, 3, 2020
|
|
6MW7
 
 | | Crystal structure of ATPase module of SMCHD1 bound to ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SODIUM ION, ... | | 著者 | Pedersen, L.C, Inoue, K, Kim, S, Perera, L, Shaw, N.D. | | 登録日 | 2018-10-29 | | 公開日 | 2019-09-11 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (2.194 Å) | | 主引用文献 | A ubiquitin-like domain is required for stabilizing the N-terminal ATPase module of human SMCHD1.
Commun Biol, 2, 2019
|
|
1HP7
 
 | | A 2.1 ANGSTROM STRUCTURE OF AN UNCLEAVED ALPHA-1-ANTITRYPSIN SHOWS VARIABILITY OF THE REACTIVE CENTER AND OTHER LOOPS | | 分子名称: | ALPHA-1-ANTITRYPSIN, BETA-MERCAPTOETHANOL, ZINC ION | | 著者 | Kim, S.-J, Woo, J.-R, Seo, E.J, Yu, M.-H, Ryu, S.-E. | | 登録日 | 2000-12-12 | | 公開日 | 2001-03-14 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A 2.1 A resolution structure of an uncleaved alpha(1)-antitrypsin shows variability of the reactive center and other loops.
J.Mol.Biol., 306, 2001
|
|
6VYP
 
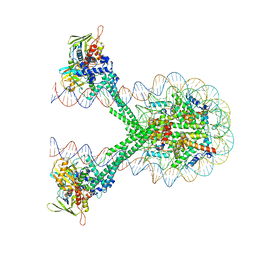 | | Crystal structure of the LSD1/CoREST histone demethylase bound to its nucleosome substrate | | 分子名称: | DNA (191-MER), FLAVIN-ADENINE DINUCLEOTIDE, Histone H2A type 1, ... | | 著者 | Kim, S, Zhu, J, Eek, P, Yennawar, N, Song, T. | | 登録日 | 2020-02-27 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (4.99 Å) | | 主引用文献 | Crystal Structure of the LSD1/CoREST Histone Demethylase Bound to Its Nucleosome Substrate.
Mol.Cell, 78, 2020
|
|
