2OG9
 
 | |
6TI7
 
 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | 分子名称: | Amyloid-beta precursor protein | | 著者 | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | 登録日 | 2019-11-21 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
6T1J
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with piperazine-urea derivative 2 | | 分子名称: | 1,2-ETHANEDIOL, Protein ENL, ~{N}-[[4-(pyrrolidin-1-ylmethyl)phenyl]methyl]-4-thiophen-2-ylcarbonyl-piperazine-1-carboxamide | | 著者 | Chaikuad, A, Heidenreich, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2019-10-04 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
2NY6
 
 | | HIV-1 gp120 Envelope Glycoprotein (M95W, W96C, I109C, T123C, T257S, V275C,S334A, S375W, Q428C, G431C) Complexed with CD4 and Antibody 17b | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, HEAVY CHAIN, ... | | 著者 | Zhou, T, Xu, L, Dey, B, Hessell, A.J, Van Ryk, D, Xiang, S.H, Yang, X, Zhang, M.Y, Zwick, M.B, Arthos, J, Burton, D.R, Dimitrov, D.S, Sodroski, J, Wyatt, R, Nabel, G.J, Kwong, P.D. | | 登録日 | 2006-11-20 | | 公開日 | 2007-02-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural definition of a conserved neutralization epitope on HIV-1 gp120.
Nature, 445, 2007
|
|
2OHF
 
 | |
3JYO
 
 | | Quinate dehydrogenase from Corynebacterium glutamicum in complex with NAD | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Quinate/shikimate dehydrogenase | | 著者 | Hoeppner, A, Niefind, K, Schomburg, D. | | 登録日 | 2009-09-22 | | 公開日 | 2010-10-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Enzyme-substrate complexes of the quinate/shikimate dehydrogenase from Corynebacterium glutamicum enable new insights in substrate and cofactor binding, specificity, and discrimination.
Biol.Chem., 394, 2013
|
|
3CFO
 
 | | Triple Mutant APO structure | | 分子名称: | DNA polymerase, GUANOSINE, SULFATE ION | | 著者 | Wang, J, Klimenko, D, Wang, M, Steitz, T.A, Konigsberg, W.H. | | 登録日 | 2008-03-04 | | 公開日 | 2009-03-10 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Insights into base selectivity from the structures
of an RB69 DNA Polymerase triple mutant
To be Published
|
|
2OIZ
 
 | |
6TAY
 
 | | Mouse RNF213 mutant R4753K modeling the Moyamoya-disease-related Human variant R4810K | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNF213,E3 ubiquitin-protein ligase RNF213,E3 ubiquitin-protein ligase RNF213, ... | | 著者 | Ahel, J, Meinhart, A, Haselbach, D, Clausen, T. | | 登録日 | 2019-10-31 | | 公開日 | 2020-07-01 | | 最終更新日 | 2021-01-13 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Moyamoya disease factor RNF213 is a giant E3 ligase with a dynein-like core and a distinct ubiquitin-transfer mechanism.
Elife, 9, 2020
|
|
2NS9
 
 | | Crystal structure of protein APE2225 from Aeropyrum pernix K1, Pfam COXG | | 分子名称: | Hypothetical protein APE2225, PHOSPHATE ION | | 著者 | Jin, X, Bera, A, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2006-11-03 | | 公開日 | 2006-11-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of hypothetical protein APE2225 from Aeropyrum pernix K1
To be Published
|
|
6T9C
 
 | |
2O0Q
 
 | | X-ray Crystal Structure of Protein CC0527 from Caulobacter crescentus. Northeast Structural Genomics Consortium Target CcR55 | | 分子名称: | Hypothetical protein CC0527 | | 著者 | Seetharaman, J, Su, M, Wang, D, Fang, Y, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-11-28 | | 公開日 | 2006-12-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of the Hypothetical Protein from Caulobacter Crescentus.
TO BE PUBLISHED
|
|
6TAC
 
 | | Human NAMPT deletion mutant in complex with nicotinamide mononucleotide, pyrophosphate, and Mg2+ | | 分子名称: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Houry, D, Raasakka, A, Kursula, P, Ziegler, M. | | 登録日 | 2019-10-29 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Identification of structural determinants of NAMPT activity and substrate selectivity
To Be Published
|
|
6TD7
 
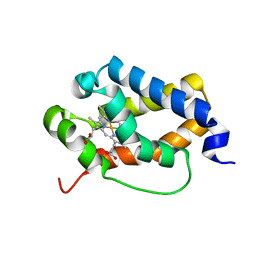 | | Structure of truncated hemoglobin THB11 from Chlamydomonas reinhardtii | | 分子名称: | CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE, THB11 | | 著者 | Huwald, D, Gasper, R, Hemschemeier, A, Hofmann, E. | | 登録日 | 2019-11-08 | | 公開日 | 2020-02-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Distinctive structural properties of THB11, a pentacoordinate Chlamydomonas reinhardtii truncated hemoglobin with N- and C-terminal extensions.
J.Biol.Inorg.Chem., 25, 2020
|
|
2O2C
 
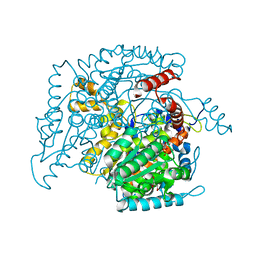 | | Crystal structure of phosphoglucose isomerase from T. brucei containing glucose-6-phosphate in the active site | | 分子名称: | GLUCOSE-6-PHOSPHATE, GLYCEROL, Glucose-6-phosphate isomerase, ... | | 著者 | Arsenieva, D, Mazock, G.H, Appavu, B.L, Jeffery, C.J. | | 登録日 | 2006-11-29 | | 公開日 | 2007-11-13 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Crystal structure of phosphoglucose isomerase from Trypanosoma brucei complexed with glucose-6-phosphate at 1.6 A resolution
Proteins, 74, 2008
|
|
3CLB
 
 | | Structure of bifunctional TcDHFR-TS in complex with TMQ | | 分子名称: | 1,2-ETHANEDIOL, DHFR-TS, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Schormann, N, Senkovich, O, Chattopadhyay, D. | | 登録日 | 2008-03-18 | | 公開日 | 2009-01-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure-based approach to pharmacophore identification, in silico screening, and three-dimensional quantitative structure-activity relationship studies for inhibitors of Trypanosoma cruzi dihydrofolate reductase function.
Proteins, 73, 2008
|
|
3CMR
 
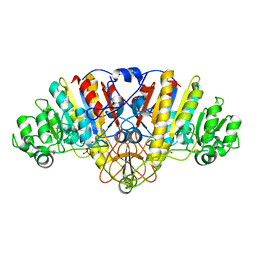 | | E. coli alkaline phosphatase mutant R166S in complex with phosphate | | 分子名称: | Alkaline phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | O'Brien, P.J, Lassila, J.K, Fenn, T.D, Zalatan, J.G, Herschlag, D. | | 登録日 | 2008-03-24 | | 公開日 | 2008-07-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Arginine coordination in enzymatic phosphoryl transfer: evaluation of the effect of Arg166 mutations in Escherichia coli alkaline phosphatase
Biochemistry, 47, 2008
|
|
3J68
 
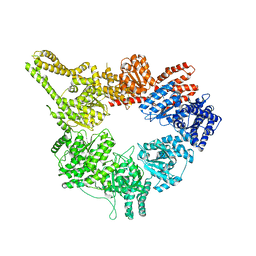 | | Structural mechanism of the dynein powerstroke (pre-powerstroke state) | | 分子名称: | Dynein motor domain | | 著者 | Lin, J, Okada, K, Raytchev, M, Smith, M.C, Nicastro, D. | | 登録日 | 2013-12-23 | | 公開日 | 2014-04-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (30 Å) | | 主引用文献 | Structural mechanism of the dynein power stroke.
Nat.Cell Biol., 16, 2014
|
|
3ZGB
 
 | | Greater efficiency of photosynthetic carbon fixation due to single amino acid substitution | | 分子名称: | 1,2-ETHANEDIOL, ASPARTIC ACID, PHOSPHOENOLPYRUVATE CARBOXYLASE, ... | | 著者 | Paulus, J.K, Schlieper, D, Groth, G. | | 登録日 | 2012-12-17 | | 公開日 | 2013-02-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Greater Efficiency of Photosynthetic Carbon Fixation due to Single Amino Acid Substitution
Nat.Commun., 4, 2013
|
|
2NWA
 
 | | X-ray Crystal Structure of Protein ytmB from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR466 | | 分子名称: | Hypothetical protein ytmB, SULFATE ION | | 著者 | Zhou, W, Forouhar, F, Seetharaman, J, Wang, D, Cunningham, K, Ma, L.-C, Fang, Y, Xiao, R, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-11-14 | | 公開日 | 2007-01-09 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal Structure of the Hypothetical protein YtmB from Bacillus subtilis subsp. (subtilis str. 168), Northeast Structural Genomics target SR466
To be Published
|
|
1FRQ
 
 | | FERREDOXIN:NADP+ OXIDOREDUCTASE (FERREDOXIN REDUCTASE) MUTANT E312A | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, PROTEIN (FERREDOXIN:NADP+ OXIDOREDUCTASE), ... | | 著者 | Aliverti, A, Deng, Z, Ravasi, D, Piubelli, L, Karplus, P.A, Zanetti, G. | | 登録日 | 1998-10-10 | | 公開日 | 1998-10-14 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Probing the function of the invariant glutamyl residue 312 in spinach ferredoxin-NADP+ reductase.
J.Biol.Chem., 273, 1998
|
|
3ZLL
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-azanyl-6-oxidanyl-1-(phenylmethyl)pyrimidine-2,4-dione, CHLORIDE ION, ... | | 著者 | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Sarkar, A, Brenk, R, Westwood, N.J, Gray, D, Naismith, J.H. | | 登録日 | 2013-02-01 | | 公開日 | 2013-02-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
6T1L
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with piperazine-urea derivative 3 | | 分子名称: | 1,2-ETHANEDIOL, Protein ENL, ~{N}-[[4-(diethylaminomethyl)phenyl]methyl]-4-pyrimidin-2-yl-piperazine-1-carboxamide | | 著者 | Chaikuad, A, Heidenreich, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2019-10-04 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
3J9G
 
 | | Atomic model of the VipA/VipB, the type six secretion system contractile sheath of Vibrio cholerae from cryo-EM | | 分子名称: | VipA, VipB | | 著者 | Kudryashev, M, Wang, R.Y.-R, Brackmann, M, Scherer, S, Maier, T, Baker, D, DiMaio, F, Stahlberg, H, Egelman, E.H, Basler, M. | | 登録日 | 2015-01-16 | | 公開日 | 2015-03-11 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of the Type VI Secretion System Contractile Sheath.
Cell(Cambridge,Mass.), 160, 2015
|
|
6T3A
 
 | | Difference-refined structure of rsEGFP2 10 ns following 400-nm laser irradiation of the off-state determined by SFX | | 分子名称: | Green fluorescent protein | | 著者 | Woodhouse, J, Coquelle, N, Adam, V, Barends, T.R.M, De La Mora, E, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | 登録日 | 2019-10-10 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Photoswitching mechanism of a fluorescent protein revealed by time-resolved crystallography and transient absorption spectroscopy.
Nat Commun, 11, 2020
|
|
