6TAM
 
 | |
6TAN
 
 | | X-RAY STRUCTURE OF HUMAN K-RAS G12C IN COMPLEX WITH COVALENT ISOQUINOLINONE INHIBITOR (COMPOUND 17) | | 分子名称: | 7-(2-fluoranyl-6-oxidanyl-phenyl)-3-[(3~{R})-1-propanoylpyrrolidin-3-yl]-4~{H}-2,6-naphthyridin-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Hillig, R.C, Eis, K, Badock, V. | | 登録日 | 2019-10-30 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Computationally Empowered Workflow Identifies Novel Covalent Allosteric Binders for KRASG12C.
Chemmedchem, 15, 2020
|
|
2QA1
 
 | | Crystal structure of PgaE, an aromatic hydroxylase involved in angucycline biosynthesis | | 分子名称: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Koskiniemi, H, Dobritzsch, D, Metsa-Ketela, M, Kallio, P, Niemi, J, Schneider, G. | | 登録日 | 2007-06-14 | | 公開日 | 2007-08-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of two aromatic hydroxylases involved in the early tailoring steps of angucycline biosynthesis
J.Mol.Biol., 372, 2007
|
|
2QA2
 
 | | Crystal structure of CabE, an aromatic hydroxylase from angucycline biosynthesis, determined to 2.7 A resolution | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Polyketide oxygenase CabE | | 著者 | Koskiniemi, H, Dobritzsch, D, Metsa-Ketela, M, Kallio, P, Niemi, J, Schneider, G. | | 登録日 | 2007-06-14 | | 公開日 | 2007-08-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structures of two aromatic hydroxylases involved in the early tailoring steps of angucycline biosynthesis
J.Mol.Biol., 372, 2007
|
|
2W70
 
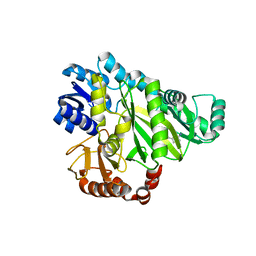 | |
2W6Q
 
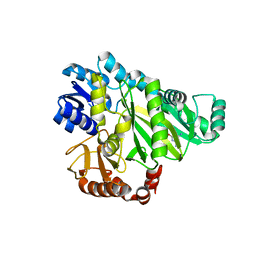 | | Crystal structure of Biotin carboxylase from E. coli in complex with the triazine-2,4-diamine fragment | | 分子名称: | 6-(2-phenoxyethoxy)-1,3,5-triazine-2,4-diamine, BIOTIN CARBOXYLASE, CHLORIDE ION | | 著者 | Mochalkin, I, Miller, J.R. | | 登録日 | 2008-12-18 | | 公開日 | 2009-05-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Discovery of Antibacterial Biotin Carboxylase Inhibitors by Virtual Screening and Fragment-Based Approaches.
Acs Chem.Biol., 4, 2009
|
|
2W71
 
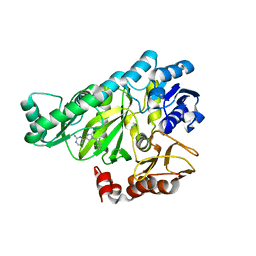 | |
4GFO
 
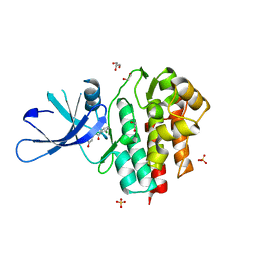 | | TYK2 kinase (JH1 domain) with 2,6-DICHLORO-N-(2-OXO-2,5-DIHYDROPYRIDIN-4-YL)BENZAMIDE | | 分子名称: | 1,2-ETHANEDIOL, 2,6-dichloro-N-(2-oxo-2,5-dihydropyridin-4-yl)benzamide, GLYCEROL, ... | | 著者 | Eigenbrot, C, Ultsch, M. | | 登録日 | 2012-08-03 | | 公開日 | 2013-06-19 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Lead identification of novel and selective TYK2 inhibitors.
Eur.J.Med.Chem., 67, 2013
|
|
4GJ3
 
 | | Tyk2 (JH1) in complex with 2,6-dichloro-4-cyano-N-[2-({[(1R,2R)-2-fluorocyclopropyl]carbonyl}amino)pyridin-4-yl]benzamide | | 分子名称: | 2,6-dichloro-4-cyano-N-[2-({[(1R,2R)-2-fluorocyclopropyl]carbonyl}amino)pyridin-4-yl]benzamide, Non-receptor tyrosine-protein kinase TYK2 | | 著者 | Ultsch, M.H. | | 登録日 | 2012-08-09 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Lead Optimization of a 4-Aminopyridine Benzamide Scaffold To Identify Potent, Selective, and Orally Bioavailable TYK2 Inhibitors.
J.Med.Chem., 56, 2013
|
|
2PU9
 
 | |
4IFH
 
 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM44619 | | 分子名称: | Insulin-degrading enzyme, N-({1-[(2R)-4-(hydroxyamino)-1-(naphthalen-2-yl)-4-oxobutan-2-yl]-1H-1,2,3-triazol-4-yl}methyl)-4-methylbenzamide, ZINC ION | | 著者 | Liang, W.G, Guo, Q, Deprez, R, Deprez, B, Tang, W. | | 登録日 | 2012-12-14 | | 公開日 | 2013-12-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.286 Å) | | 主引用文献 | Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice.
Nat Commun, 6, 2015
|
|
5N9C
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-PP-[ProM-1]-OH | | 分子名称: | Ac-[2-Cl-F]-PP-[ProM-1]-OH, GLYCEROL, NITRATE ION, ... | | 著者 | Barone, M, Roske, Y. | | 登録日 | 2017-02-24 | | 公開日 | 2017-06-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4I2Y
 
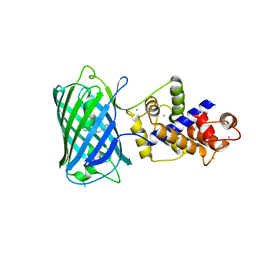 | |
2F74
 
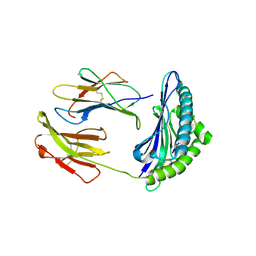 | | Murine MHC class I H-2Db in complex with human b2-microglobulin and LCMV-derived immunodminant peptide gp33 | | 分子名称: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | 著者 | Achour, A, Michaelsson, J, Harris, R.A, Ljunggren, H.G, Karre, K, Schneider, G, Sandalova, T. | | 登録日 | 2005-11-30 | | 公開日 | 2006-02-14 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Basis of the Differential Stability and Receptor Specificity of H-2D(b) in Complex with Murine versus Human beta(2)-Microglobulin.
J.Mol.Biol., 356, 2006
|
|
5NCF
 
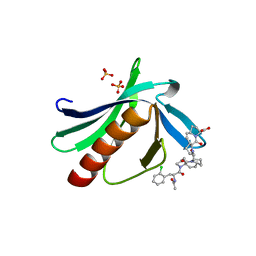 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-4]-OH | | 分子名称: | (1~{S},4~{S},7~{R},10~{R})-14-[(3~{S},6~{R},8~{a}~{S})-1'-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8~{a}-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonyl-2-oxidanylidene-3,14-diazatricyclo[8.4.0.0^{3,7}]tetradec-8-ene-4-carboxylic acid, GLYCEROL, NITRATE ION, ... | | 著者 | Barone, M, Roske, Y. | | 登録日 | 2017-03-04 | | 公開日 | 2017-06-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5N9P
 
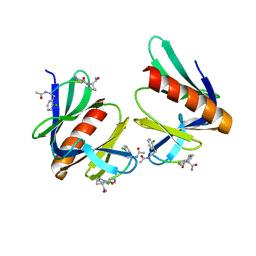 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-PP-[ProM-1]-NH2 | | 分子名称: | Ac-[2-Cl-F]-PP-[ProM-1]-NH2, CHLORIDE ION, Protein enabled homolog | | 著者 | Barone, M, Roske, Y. | | 登録日 | 2017-02-26 | | 公開日 | 2017-06-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1NMF
 
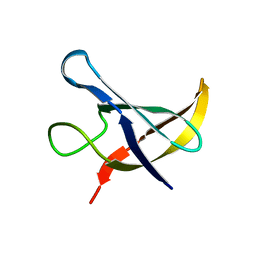 | |
1NMG
 
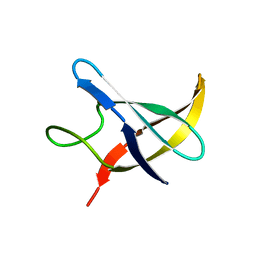 | |
3U0L
 
 | | Crystal structure of the engineered fluorescent protein mRuby, crystal form 1, pH 4.5 | | 分子名称: | ACETATE ION, mRuby | | 著者 | Akerboom, J, Looger, L.L, Schreiter, E.R. | | 登録日 | 2011-09-28 | | 公開日 | 2012-10-03 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics.
Front Mol Neurosci, 6, 2013
|
|
3U0M
 
 | |
5LN3
 
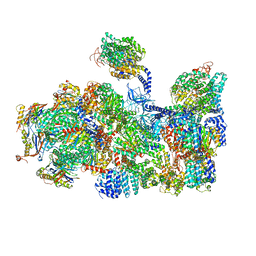 | | The human 26S Proteasome at 6.8 Ang. | | 分子名称: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | 著者 | Schweitzer, A, Beck, F, Sakata, E, Unverdorben, P. | | 登録日 | 2016-08-03 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (6.8 Å) | | 主引用文献 | Molecular Details Underlying Dynamic Structures and Regulation of the Human 26S Proteasome.
Mol. Cell Proteomics, 16, 2017
|
|
7Z1A
 
 | | Nanobody H11 and F2 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2 Nanobody, H11 Nanobody, ... | | 著者 | Mikolajek, H, Naismith, J.H. | | 登録日 | 2022-02-24 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1D
 
 | | Nanobody H11-H6 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, H11-H6 nanobody, ... | | 著者 | Mikolajek, H, Naismith, J.H. | | 登録日 | 2022-02-24 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1C
 
 | | Nanobody H11-B5 and H11-F2 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Nanobody B5, ... | | 著者 | Mikolajek, H, Naismith, J.H. | | 登録日 | 2022-02-24 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1B
 
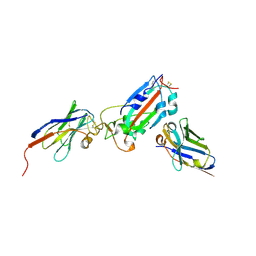 | | Nanobody H11-A10 and F2 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody A10, Nanobody F2, ... | | 著者 | Mikolajek, H, Naismith, J.H. | | 登録日 | 2022-02-24 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
