8B23
 
 | |
8B21
 
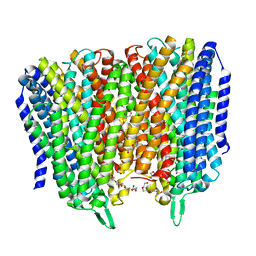 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 0-60-seconds post reaction initiation with Na+ | | 分子名称: | DI(HYDROXYETHYL)ETHER, DODECYL-BETA-D-MALTOSIDE, K(+)-stimulated pyrophosphate-energized sodium pump, ... | | 著者 | Strauss, J, Vidilaseris, K, Goldman, A. | | 登録日 | 2022-09-12 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
5HLL
 
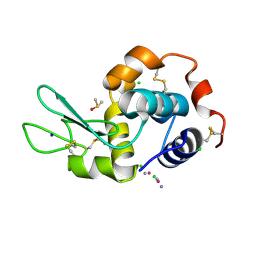 | |
5I6U
 
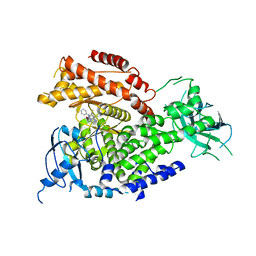 | | The crystal structure of PI3Kdelta with compound 32 | | 分子名称: | 2-[(1S)-1-({6-amino-5-[(1H-pyrazol-4-yl)ethynyl]pyrimidin-4-yl}amino)ethyl]-5-chloro-3-phenylquinazolin-4(3H)-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | 著者 | Somoza, J.R, Villasenor, A.G. | | 登録日 | 2016-02-16 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.842 Å) | | 主引用文献 | The crystal structure of PI3Kdelta with compound 32
To Be Published
|
|
5MJG
 
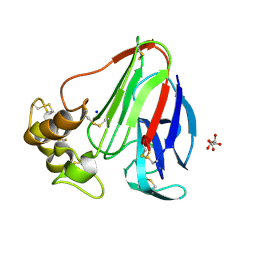 | | Single-shot pink beam serial crystallography: Thaumatin | | 分子名称: | S,R MESO-TARTARIC ACID, SODIUM ION, Thaumatin-1 | | 著者 | Meents, A, Oberthuer, D, Lieske, J, Srajer, V. | | 登録日 | 2016-12-01 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Single-shot pink beam serial crystallography: Thaumatin
To Be Published
|
|
7UK2
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with NN-390 | | 分子名称: | Hdac6 protein, N-hydroxy-4-{[(propan-2-yl)(2,3,4,5-tetrafluorobenzene-1-sulfonyl)amino]methyl}benzamide, POTASSIUM ION, ... | | 著者 | Erdogan, F, Seo, H.-S, Dhe-Paganon, S. | | 登録日 | 2022-03-31 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | High Efficacy and Drug Synergy of HDAC6-Selective Inhibitor NN-429 in Natural Killer (NK)/T-Cell Lymphoma.
Pharmaceuticals, 15, 2022
|
|
6ZF5
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | 分子名称: | 1-[3-[(4-butylphenyl)sulfonyl-(2-hydroxy-2-oxoethyl)amino]phenyl]-5-cyclopropyl-pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | 著者 | Narayanan, D, Bach, A, Gajhede, M. | | 登録日 | 2020-06-16 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZEX
 
 | | Keap1 kelch domain bound to a small molecule fragment | | 分子名称: | 5-cyclopropyl-1-phenyl-pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1 | | 著者 | Narayanan, D, Bach, A, Gajhede, M. | | 登録日 | 2020-06-16 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
8SCA
 
 | | Rec3 Domain from S. pyogenes Cas9 | | 分子名称: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9/Csn1 | | 著者 | D'Ordine, A.M, Skeens, E, Lisi, G.P, Jogl, G. | | 登録日 | 2023-04-05 | | 公開日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | High-fidelity, hyper-accurate, and evolved mutants rewire atomic-level communication in CRISPR-Cas9.
Sci Adv, 10, 2024
|
|
8DTA
 
 | |
8EE2
 
 | |
8E2N
 
 | |
6XR3
 
 | |
7TN1
 
 | | Multistate design to stabilize viral class I fusion proteins | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0 | | 著者 | Huang, J, Banerjee, A, Gonzalez, K, Mousa, J, Strauch, E. | | 登録日 | 2022-01-20 | | 公開日 | 2023-07-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Multistate design to stabilize viral class I fusion proteins
To Be Published
|
|
6Z2H
 
 | |
8TJG
 
 | |
7AYG
 
 | | oxalyl-CoA decarboxylase from Methylorubrum extorquens with bound TPP and ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative oxalyl-CoA decarboxylase (Oxc, ... | | 著者 | Pfister, P, Burgener, S, Nattermann, M, Zarzycki, J, Erb, T.J. | | 登録日 | 2020-11-12 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Engineering a Highly Efficient Carboligase for Synthetic One-Carbon Metabolism.
Acs Catalysis, 11, 2021
|
|
7B2E
 
 | | quadruple mutant of oxalyl-CoA decarboxylase from Methylorubrum extorquens with bound TPP and ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative oxalyl-CoA decarboxylase (Oxc, ... | | 著者 | Pfister, P, Burgener, S, Nattermann, M, Zarzycki, J, Erb, T.J. | | 登録日 | 2020-11-26 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Engineering a Highly Efficient Carboligase for Synthetic One-Carbon Metabolism.
Acs Catalysis, 11, 2021
|
|
6YLV
 
 | | Translation initiation factor 4E in complex with 4-Cl-Bn7GpppG mRNA 5' cap analog | | 分子名称: | 4-Cl-Bn7GpppG mRNA 5' cap analog, Eukaryotic translation initiation factor 4E, GLYCEROL | | 著者 | Kubacka, D, Wojcik, R, Baranowski, M.R, Kowalska, J, Jemielity, J. | | 登録日 | 2020-04-07 | | 公開日 | 2020-06-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.66005659 Å) | | 主引用文献 | Novel N7-Arylmethyl Substituted Dinucleotide mRNA 5' cap Analogs: Synthesis and Evaluation as Modulators of Translation.
Pharmaceutics, 13, 2021
|
|
6XTV
 
 | | FULL-LENGTH LTTR LYSG FROM CORYNEBACTERIUM GLUTAMICUM WITH BOUND EFFECTOR ARG | | 分子名称: | ARGININE, Lysine export transcriptional regulatory protein LysG | | 著者 | Hofmann, E, Syberg, F, Schlicker, C, Eggeling, L, Schendzielorz, G. | | 登録日 | 2020-01-16 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Engineering and application of a biosensor with focused ligand specificity.
Nat Commun, 11, 2020
|
|
6T6H
 
 | |
6T6Z
 
 | |
6T6Y
 
 | |
6T6X
 
 | | Structure of the Bottromycin epimerase BotH in complex with substrate | | 分子名称: | (4~{R})-2-[(1~{R})-1-[[(2~{S})-2-[[(2~{S})-3-methyl-2-[[(4~{Z},6~{S},9~{S},12~{S})-2,8,11-tris(oxidanylidene)-6,9-di(propan-2-yl)-1,4,7,10-tetrazabicyclo[10.3.0]pentadec-4-en-5-yl]amino]butanoyl]amino]-3-phenyl-propanoyl]amino]-3-oxidanyl-3-oxidanylidene-propyl]-4,5-dihydro-1,3-thiazole-4-carboxylic acid, BotH | | 著者 | Koehnke, J, Sikandar, A. | | 登録日 | 2019-10-20 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | The bottromycin epimerase BotH defines a group of atypical alpha / beta-hydrolase-fold enzymes.
Nat.Chem.Biol., 16, 2020
|
|
6T70
 
 | |
