5X0W
 
 | | Molecular mechanism for the binding between Sharpin and HOIP | | 分子名称: | E3 ubiquitin-protein ligase RNF31, Sharpin | | 著者 | Liu, J, Li, F, Cheng, X, Pan, L. | | 登録日 | 2017-01-23 | | 公開日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Insights into SHARPIN-Mediated Activation of HOIP for the Linear Ubiquitin Chain Assembly
Cell Rep, 21, 2017
|
|
4FZA
 
 | | Crystal structure of MST4-MO25 complex | | 分子名称: | Calcium-binding protein 39, GLYCEROL, Serine/threonine-protein kinase MST4 | | 著者 | Shi, Z.B, Zhou, Z.C. | | 登録日 | 2012-07-06 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
4FZF
 
 | | Crystal structure of MST4-MO25 complex with DKI | | 分子名称: | 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, Calcium-binding protein 39, Serine/threonine-protein kinase MST4 | | 著者 | Shi, Z.B, Zhou, Z.C. | | 登録日 | 2012-07-06 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.64 Å) | | 主引用文献 | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
4FZD
 
 | | Crystal structure of MST4-MO25 complex with WSF motif | | 分子名称: | C-terminal peptide from Serine/threonine-protein kinase MST4, Calcium-binding protein 39, GLYCEROL, ... | | 著者 | Shi, Z.B, Zhou, Z.C. | | 登録日 | 2012-07-06 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
4X2S
 
 | |
4WNX
 
 | | Netrin 4 lacking the C-terminal Domain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | McDougall, M, Patel, T, Reuten, R, Meier, M, Koch, M, Stetefeld, J. | | 登録日 | 2014-10-14 | | 公開日 | 2016-02-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.723 Å) | | 主引用文献 | Structural decoding of netrin-4 reveals a regulatory function towards mature basement membranes.
Nat Commun, 7, 2016
|
|
2H5E
 
 | |
4GEH
 
 | |
4I1L
 
 | | Structural and Biological Features of FOXP3 Dimerization Relevant to Regulatory T Cell Function | | 分子名称: | ACETATE ION, Forkhead box protein P3, MAGNESIUM ION, ... | | 著者 | Song, X.M, Greene, M.I, Zhou, Z.C. | | 登録日 | 2012-11-21 | | 公開日 | 2012-12-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and biological features of FOXP3 dimerization relevant to regulatory T cell function.
Cell Rep, 1, 2012
|
|
4KQE
 
 | | The mutant structure of the human glycyl-tRNA synthetase E71G | | 分子名称: | GLYCEROL, Glycine--tRNA ligase | | 著者 | Qin, X, Hao, Z, Tian, Q, Zhang, Z, Zhou, C, Xie, W. | | 登録日 | 2013-05-15 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.739 Å) | | 主引用文献 | Large Conformational Changes of Insertion 3 in Human Glycyl-tRNA Synthetase (hGlyRS) during Catalysis
J.Biol.Chem., 291, 2016
|
|
6M3M
 
 | |
8XS5
 
 | |
8XNG
 
 | |
8XW4
 
 | |
8XW0
 
 | | Cryo-EM structure of OSCA3.1-GDN state | | 分子名称: | CSC1-like protein ERD4, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine | | 著者 | Zhang, Y, Han, Y. | | 登録日 | 2024-01-15 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XW3
 
 | |
8XRY
 
 | |
8XW2
 
 | |
8XVZ
 
 | |
6M4G
 
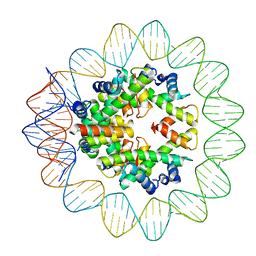 | | Structural mechanism of nucleosome dynamics governed by human histone variants H2A.B and H2A.Z.2.2 | | 分子名称: | DNA (93-MER), Histone H2A-Bbd type 2/3, Histone H2B type 2-E, ... | | 著者 | Zhou, M, Dai, L.C, Li, C.M, Shi, L.X, Huang, Y, Guo, Z.Q. | | 登録日 | 2020-03-06 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis of nucleosome dynamics modulation by histone variants H2A.B and H2A.Z.2.2.
Embo J., 40, 2021
|
|
8XAJ
 
 | |
8XW1
 
 | |
8XVX
 
 | |
8XS0
 
 | |
8XS4
 
 | |
