5YJH
 
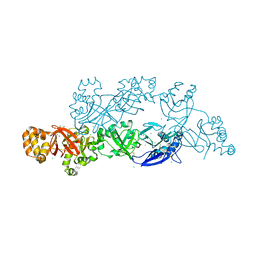 | | Structural insights into periostin functions | | 分子名称: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Liu, H, Liu, J, Xu, F. | | 登録日 | 2017-10-10 | | 公開日 | 2018-05-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.957 Å) | | 主引用文献 | Structural characterizations of human periostin dimerization and cysteinylation.
FEBS Lett., 592, 2018
|
|
5YJG
 
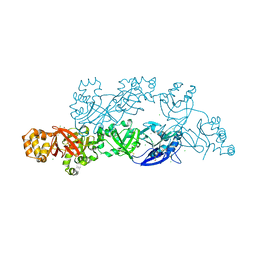 | | Structural insights into periostin functions | | 分子名称: | CALCIUM ION, CHLORIDE ION, CYSTEINE, ... | | 著者 | Liu, H, Liu, J, Xu, F. | | 登録日 | 2017-10-10 | | 公開日 | 2018-05-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.399 Å) | | 主引用文献 | Structural characterizations of human periostin dimerization and cysteinylation.
FEBS Lett., 592, 2018
|
|
5ZVS
 
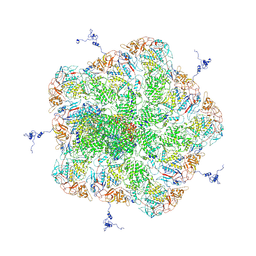 | |
5XGP
 
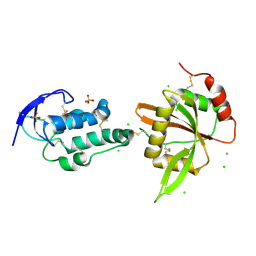 | |
5ZVT
 
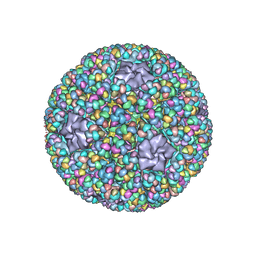 | | Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly | | 分子名称: | C-terminus of outer capsid protein VP5, Core protein VP6, MYRISTIC ACID, ... | | 著者 | Liu, H, Fang, Q, Cheng, L. | | 登録日 | 2018-05-12 | | 公開日 | 2018-07-04 | | 最終更新日 | 2018-07-25 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5Y4M
 
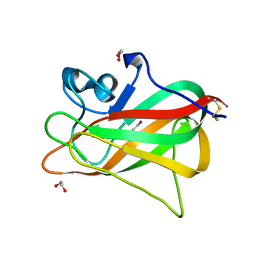 | | Discoidin domain of human CASPR2 | | 分子名称: | 1,2-ETHANEDIOL, human CASPR2 Disc domain | | 著者 | Liu, H, Xu, F, Zhang, J, Liang, W. | | 登録日 | 2017-08-04 | | 公開日 | 2018-08-08 | | 最終更新日 | 2019-02-20 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Structural mapping of hot spots within human CASPR2 discoidin domain for autoantibody recognition.
J. Autoimmun., 96, 2019
|
|
6AX3
 
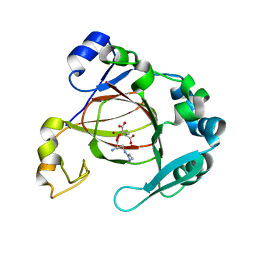 | | Complex structure of JMJD5 and Symmetric Dimethyl-Arginine (SDMA) | | 分子名称: | 2-OXOGLUTARIC ACID, Lysine-specific demethylase 8, N3, ... | | 著者 | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | 登録日 | 2017-09-06 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
6AVS
 
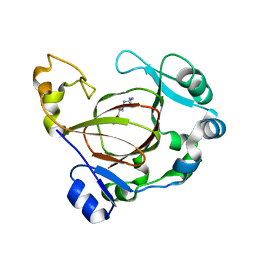 | | Complex structure of JMJD5 and Symmetric Monomethyl-Arginine (MMA) | | 分子名称: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, Lysine-specific demethylase 8, ZINC ION | | 著者 | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | 登録日 | 2017-09-04 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
8JOV
 
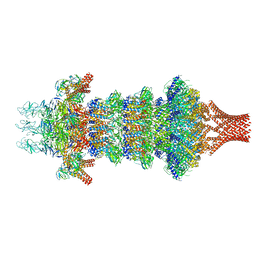 | | Portal-tail complex of phage GP4 | | 分子名称: | Portal protein, Putative tail fiber protein, Virion associated protein, ... | | 著者 | Liu, H, Chen, W. | | 登録日 | 2023-06-08 | | 公開日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Asymmetric Structure of Podophage GP4 Reveals a Novel Architecture of Three Types of Tail Fibers.
J.Mol.Biol., 435, 2023
|
|
8JOU
 
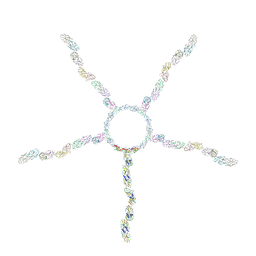 | | Fiber I and fiber-tail-adaptor of phage GP4 | | 分子名称: | Virion-associated phage protein, rope protein of phage GP4 | | 著者 | Liu, H, Chen, W. | | 登録日 | 2023-06-08 | | 公開日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Asymmetric Structure of Podophage GP4 Reveals a Novel Architecture of Three Types of Tail Fibers.
J.Mol.Biol., 435, 2023
|
|
8I4L
 
 | | Capsid structure of the Cyanophage P-SCSP1u | | 分子名称: | The capsid protein(gp 19) of P-SCSP1u | | 著者 | Liu, H, Dang, S. | | 登録日 | 2023-01-19 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | Cryo-EM structure of cyanophage P-SCSP1u offers insights into DNA gating and evolution of T7-like viruses.
Nat Commun, 14, 2023
|
|
8I4M
 
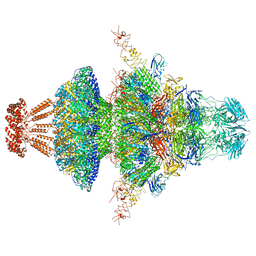 | | Portal-tail complex structure of the Cyanophage P-SCSP1u | | 分子名称: | Adaptor protein(gp22) of the cyanophage P-SCSP1u, Fiber protein(gp 28) of the cyanophage P-SCSP1u, Nozzle protein(gp 23) of the cyanophage P-SCSP1u, ... | | 著者 | Liu, H, Dang, S. | | 登録日 | 2023-01-19 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.81 Å) | | 主引用文献 | Cryo-EM structure of cyanophage P-SCSP1u offers insights into DNA gating and evolution of T7-like viruses.
Nat Commun, 14, 2023
|
|
8JP2
 
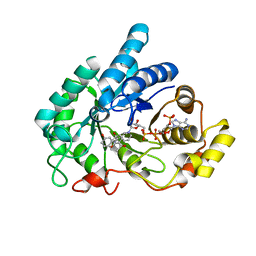 | | Crystal structure of AKR1C1 in complex with DFV | | 分子名称: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zheng, X.H, Liu, H, Yao, Z.Q, Zhang, L.P. | | 登録日 | 2023-06-10 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Inhibition of AKR1Cs by liquiritigenin and the structural basis.
Chem.Biol.Interact., 385, 2023
|
|
8JP1
 
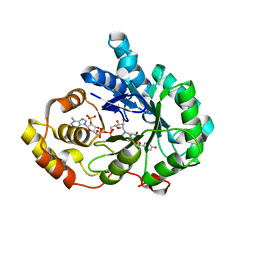 | | Crystal structure of AKR1C3 in complex with DFV | | 分子名称: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zheng, X.H, Liu, H, Yao, Z.Q, Zhang, L.P. | | 登録日 | 2023-06-10 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Inhibition of AKR1Cs by liquiritigenin and the structural basis.
Chem.Biol.Interact., 385, 2023
|
|
7U4L
 
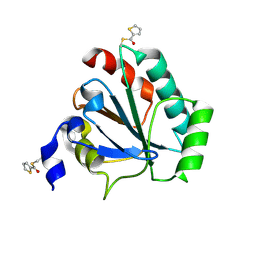 | | Crystal structure of human GPX4-U46C in complex with MAC-5576 | | 分子名称: | Phospholipid hydroperoxide glutathione peroxidase, thiophene-2-carbaldehyde | | 著者 | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | 登録日 | 2022-02-28 | | 公開日 | 2022-12-07 | | 最終更新日 | 2022-12-28 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4K
 
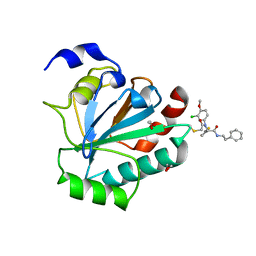 | | Crystal structure of human GPX4-U46C-R152H in complex with ML162 | | 分子名称: | 1,2-ETHANEDIOL, 2-chloro-N-(3-chloro-4-methoxyphenyl)-N-[(1R)-2-oxo-2-[(2-phenylethyl)amino]-1-(thiophen-2-yl)ethyl]acetamide, Phospholipid hydroperoxide glutathione peroxidase | | 著者 | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Xia, X, Soni, R.K, Stockwell, B.R. | | 登録日 | 2022-02-28 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4N
 
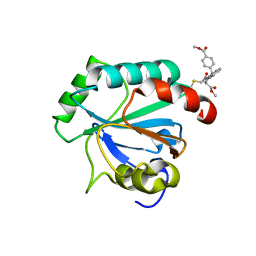 | | Crystal structure of human GPX4-U46C in complex with RSL3 | | 分子名称: | Phospholipid hydroperoxide glutathione peroxidase, methyl (1S,3R)-2-(chloroacetyl)-1-[4-(methoxycarbonyl)phenyl]-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxylate | | 著者 | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Xia, X, Soni, R.K, Stockwell, B.R. | | 登録日 | 2022-02-28 | | 公開日 | 2022-12-07 | | 最終更新日 | 2022-12-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4M
 
 | | Crystal structure of human GPX4-U46C in complex with LOC1886 | | 分子名称: | 1,2-ETHANEDIOL, 4-methoxy-1H-indole-2-carbaldehyde, Phospholipid hydroperoxide glutathione peroxidase | | 著者 | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | 登録日 | 2022-02-28 | | 公開日 | 2022-12-07 | | 最終更新日 | 2022-12-28 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4J
 
 | | Crystal structure of human GPX4-U46C-R152H in complex with TMT10 | | 分子名称: | Phospholipid hydroperoxide glutathione peroxidase, THIOCYANATE ION, ~{N}-(3-chloranyl-4-methoxy-phenyl)ethanamide | | 著者 | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | 登録日 | 2022-02-28 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4I
 
 | | Crystal structure of human GPX4-U46C-R152H in complex with CDS9 | | 分子名称: | 2-bromo-N-[(thiophen-2-yl)methyl]acetamide, Phospholipid hydroperoxide glutathione peroxidase, THIOCYANATE ION | | 著者 | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | 登録日 | 2022-02-28 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
2XU2
 
 | | Crystal Structure of the hypothetical protein PA4511 from Pseudomonas aeruginosa | | 分子名称: | CITRIC ACID, UPF0271 PROTEIN PA4511 | | 著者 | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, McMahon, S.A, White, M.F, Naismith, J.H. | | 登録日 | 2010-10-14 | | 公開日 | 2011-01-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
4UX5
 
 | | Structure of DNA complex of PCG2 | | 分子名称: | 5'-D(*CP*AP*AP*TP*GP*AP*CP*GP*CP*GP*TP*AP*AP*GP)-3', 5'-D(*CP*TP*TP*AP*CP*GP*CP*GP*TP*CP*AP*TP*TP*GP)-3', TRANSCRIPTION FACTOR MBP1 | | 著者 | Liu, J, Huang, J, Zhao, Y, Liu, H, Wang, D, Yang, J, Zhao, W, Taylor, I.A, Peng, Y. | | 登録日 | 2014-08-19 | | 公開日 | 2015-01-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Basis of DNA Recognition by Pcg2 Reveals a Novel DNA Binding Mode for Winged Helix-Turn-Helix Domains.
Nucleic Acids Res., 43, 2015
|
|
8ZJD
 
 | | Cryo-EM structure of kisspeptin receptor bound to KP-10 | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1,Guanine nucleotide-binding protein G(q) subunit alpha, ... | | 著者 | Shen, S, Liu, H, Xu, H.E. | | 登録日 | 2024-05-14 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Cryo-EM structure of kisspeptin receptor bound to KP-10
To Be Published
|
|
8ZJE
 
 | | Cryo-EM structure of kisspeptin receptor bound to TAK-448 | | 分子名称: | ACY-DTY-HYP-ASN-THR-PHE-XZA-LEU-NMM-TRP-NH2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Shen, S, Liu, H, Xu, H.E. | | 登録日 | 2024-05-14 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Cryo-EM structure of kisspeptin receptor bound to TAK-448
To Be Published
|
|
6DDA
 
 | | Nurr1 Covalently Modified by a Dopamine Metabolite | | 分子名称: | 5-hydroxy-1,2-dihydro-6H-indol-6-one, BROMIDE ION, Nuclear receptor subfamily 4 group A member 2, ... | | 著者 | Bruning, J.M, Wang, Y, Otrabella, F, Boxue, T, Liu, H, Bhattacharya, P, Guo, S, Holton, J.M, Fletterick, R.J, Jacobson, M.P, England, P.M. | | 登録日 | 2018-05-09 | | 公開日 | 2019-03-20 | | 最終更新日 | 2019-11-06 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Covalent Modification and Regulation of the Nuclear Receptor Nurr1 by a Dopamine Metabolite.
Cell Chem Biol, 26, 2019
|
|
