5W5N
 
 | | Crystal structure of human IgG4-Sigma2 Fc fragment | | 分子名称: | ACETATE ION, GLYCEROL, Immunoglobulin heavy constant gamma 4, ... | | 著者 | Armstrong, A.A, Gilliland, G.L. | | 登録日 | 2017-06-15 | | 公開日 | 2017-09-13 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Functional, Biophysical, and Structural Characterization of Human IgG1 and IgG4 Fc Variants with Ablated Immune Functionality.
Antibodies, 6, 2017
|
|
5W5M
 
 | | Crystal structure of human IgG4-Sigma1 Fc fragment | | 分子名称: | ACETATE ION, GLYCEROL, Immunoglobulin heavy constant gamma 4, ... | | 著者 | Armstrong, A.A, Gilliland, G.L. | | 登録日 | 2017-06-15 | | 公開日 | 2017-09-13 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Functional, Biophysical, and Structural Characterization of Human IgG1 and IgG4 Fc Variants with Ablated Immune Functionality.
Antibodies, 6, 2017
|
|
5Z43
 
 | | Crystal structure of prenyltransferase AmbP1 apo structure | | 分子名称: | AmbP1, MAGNESIUM ION | | 著者 | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | 登録日 | 2018-01-10 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.361 Å) | | 主引用文献 | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z45
 
 | | Crystal structure of prenyltransferase AmbP1 pH6.5 complexed with GSPP and cis-indolyl vinyl isonitrile | | 分子名称: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, AmbP1, GERANYL S-THIOLODIPHOSPHATE, ... | | 著者 | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | 登録日 | 2018-01-10 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.601 Å) | | 主引用文献 | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5WQ1
 
 | |
5VSB
 
 | | Structure of DUB complex | | 分子名称: | 7-chloro-3-{[4-hydroxy-1-(3-phenylpropanoyl)piperidin-4-yl]methyl}quinazolin-4(3H)-one, Ubiquitin carboxyl-terminal hydrolase 7 | | 著者 | Seo, H.-S, Dhe-Paganon, S. | | 登録日 | 2017-05-11 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7.
Cell Chem Biol, 24, 2017
|
|
5VS6
 
 | | Structure of DUB complex | | 分子名称: | ACETATE ION, GLYCEROL, N-[3-({4-hydroxy-1-[(3R)-3-phenylbutanoyl]piperidin-4-yl}methyl)-4-oxo-3,4-dihydroquinazolin-7-yl]-3-(4-methylpiperazin-1-yl)propanamide, ... | | 著者 | Seo, H.-S, Dhe-Paganon, S. | | 登録日 | 2017-05-11 | | 公開日 | 2017-12-20 | | 最終更新日 | 2018-01-03 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7.
Cell Chem Biol, 24, 2017
|
|
5W5L
 
 | | Crystal structure of human IgG1-Sigma Fc fragment | | 分子名称: | Immunoglobulin gamma-1 heavy chain, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Armstrong, A.A, Gilliland, G.L. | | 登録日 | 2017-06-15 | | 公開日 | 2017-09-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Functional, Biophysical, and Structural Characterization of Human IgG1 and IgG4 Fc Variants with Ablated Immune Functionality.
Antibodies, 6, 2017
|
|
5VSK
 
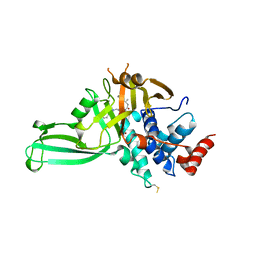 | | Structure of DUB complex | | 分子名称: | 7-chloro-3-({4-hydroxy-1-[(3S)-3-phenylbutanoyl]piperidin-4-yl}methyl)quinazolin-4(3H)-one, Ubiquitin carboxyl-terminal hydrolase 7, ZINC ION | | 著者 | Seo, H.-Y, Dhe-Paganon, S. | | 登録日 | 2017-05-11 | | 公開日 | 2017-12-20 | | 最終更新日 | 2018-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.33 Å) | | 主引用文献 | Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7.
Cell Chem Biol, 24, 2017
|
|
7XUX
 
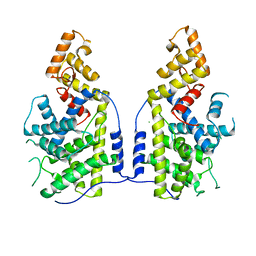 | |
5Z44
 
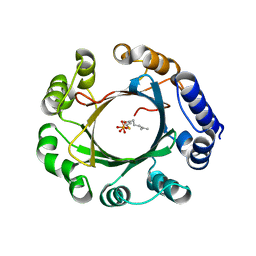 | | Crystal structure of prenyltransferase AmbP1 complexed with GSPP | | 分子名称: | AmbP1, GERANYL S-THIOLODIPHOSPHATE, MAGNESIUM ION | | 著者 | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | 登録日 | 2018-01-10 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.458 Å) | | 主引用文献 | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z46
 
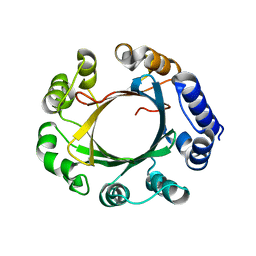 | | Crystal structure of prenyltransferase AmbP1 pH8 complexed with GSPP and cis-indolyl vinyl isonitrile | | 分子名称: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, AmbP1, GERANYL S-THIOLODIPHOSPHATE, ... | | 著者 | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | 登録日 | 2018-01-10 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7VC8
 
 | | Complex structure of AtHPPD with inhibitor PYQ3 | | 分子名称: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, pyren-1-yl 2-[1,5-dimethyl-2,4-bis(oxidanylidene)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazolin-3-yl]ethanoate | | 著者 | Yang, G.F, Lin, H.Y. | | 登録日 | 2021-09-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.606 Å) | | 主引用文献 | Design of an HPPD fluorescent probe and visualization of plant responses to abiotic stress
Adv Agrochem, 2022
|
|
7VUN
 
 | | Design, modification, evaluation and cocrystal studies of novel phthalimides regulating PD-1/PD-L1 interaction | | 分子名称: | (2~{S},3~{S})-2-[[6-[(3-cyanophenyl)methoxy]-2-(2-methyl-3-phenyl-phenyl)-1,3-bis(oxidanylidene)isoindol-5-yl]methylamino]-3-oxidanyl-butanoic acid, Programmed cell death 1 ligand 1 | | 著者 | Cheng, Y, Sun, C.L, Chen, M.R, Yang, P, Xiao, Y.B. | | 登録日 | 2021-11-03 | | 公開日 | 2022-09-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | Novel phthalimides regulating PD-1/PD-L1 interaction as potential immunotherapy agents.
Acta Pharm Sin B, 12, 2022
|
|
7W1S
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain in complex with neutralizing nanobody Nb-007 | | 分子名称: | Nanobody Nb-007, Spike protein S1 | | 著者 | Yang, J, Lin, S, Sun, H.L, Lu, G.W. | | 登録日 | 2021-11-20 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.997 Å) | | 主引用文献 | A Potent Neutralizing Nanobody Targeting the Spike Receptor-Binding Domain of SARS-CoV-2 and the Structural Basis of Its Intimate Binding.
Front Immunol, 13, 2022
|
|
7WJ8
 
 | | Complex structure of AtHPPD-PyQ1 | | 分子名称: | 2-pyren-1-yloxyethyl 2-[1,5-dimethyl-2,4-bis(oxidanylidene)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazolin-3-yl]ethanoate, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | 著者 | Yang, G.-F, Lin, H.-Y, Dong, J. | | 登録日 | 2022-01-05 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.805 Å) | | 主引用文献 | Design of an HPPD fluorescent probe and visualization of plant responses to abiotic stress
Adv Agrochem, 2022
|
|
7WJJ
 
 | | Complex structure of AtHPPD-PyQ2 | | 分子名称: | 2-[1,5-dimethyl-2,4-bis(oxidanylidene)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazolin-3-yl]-N-(2-pyren-1-yloxyethyl)ethanamide, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | 著者 | Yang, G.-F, Lin, H.-Y, Dong, J. | | 登録日 | 2022-01-06 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.603 Å) | | 主引用文献 | Design of an HPPD fluorescent probe and visualization of plant responses to abiotic stress
Adv Agrochem, 2022
|
|
7WVR
 
 | |
4YK9
 
 | |
7XE8
 
 | |
7XR5
 
 | | Crystal structure of imine reductase with NAPDH from Streptomyces albidoflavus | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39,42,45,48,51,54,57-nonadecaoxanonapentacontane-1,59-diol, 6-phosphogluconate dehydrogenase NAD-binding, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zhang, J, Chen, R.C, Gao, S.S. | | 登録日 | 2022-05-09 | | 公開日 | 2022-10-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Actinomycetes-derived imine reductases with a preference towards bulky amine substrates.
Commun Chem, 5, 2022
|
|
7X4B
 
 | | Crystal Structure of An Anti-CRISPR Protein | | 分子名称: | Anti-CRISPR protein (AcrIIC1), SULFATE ION | | 著者 | Hu, J, Zhang, S, Gao, J.Y, Liu, X, Liu, J. | | 登録日 | 2022-03-02 | | 公開日 | 2022-10-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
7XVN
 
 | |
8JMT
 
 | | Structure of the adhesion GPCR ADGRL3 in the apo state | | 分子名称: | Adhesion G protein-coupled receptor L3,Soluble cytochrome b562 | | 著者 | Tao, Y, Guo, Q, He, B, Zhong, Y. | | 登録日 | 2023-06-05 | | 公開日 | 2023-09-06 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.36 Å) | | 主引用文献 | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8KIF
 
 | | The structure of MmaE with substrate | | 分子名称: | (3R)-3-(2-hydroxy-2-oxoethylamino)decanoic acid, FE (II) ION, Putative dioxygenase | | 著者 | Chen, J, Zhou, J. | | 登録日 | 2023-08-23 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Variation in biosynthesis and metal-binding properties of isonitrile-containing peptides produced by Mycobacteria versus Streptomyces.
Acs Catalysis, 14, 2024
|
|
