6AWK
 
 | |
6MIM
 
 | |
7KTP
 
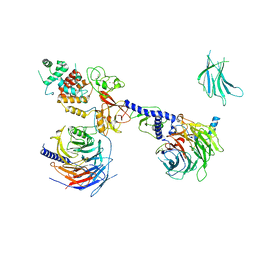 | | PRC2:EZH1_B from a dimeric PRC2 bound to a nucleosome | | 分子名称: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH1, Polycomb protein EED, ... | | 著者 | Grau, D.J, Armache, K.J. | | 登録日 | 2020-11-24 | | 公開日 | 2021-02-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction.
Nat Commun, 12, 2021
|
|
6MIO
 
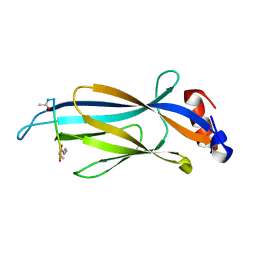 | | Crystal structure of Taf14 YEATS domain in complex with histone H3K9pr | | 分子名称: | Histone H3K9pr, Transcription initiation factor TFIID subunit 14 | | 著者 | Klein, B.J, Andrews, F.H, Vann, K.R, Kutateladze, T.G. | | 登録日 | 2018-09-19 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
6AZG
 
 | |
6MIQ
 
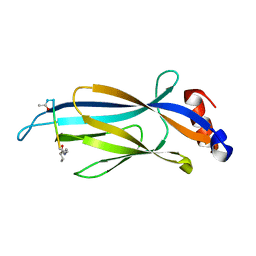 | | Crystal structure of Taf14 YEATS domain in complex with histone H3K9bu | | 分子名称: | Histone H3K9bu, Transcription initiation factor TFIID subunit 14 | | 著者 | Klein, B.J, Andrews, F.H, Vann, K.R, Kutateladze, T.G. | | 登録日 | 2018-09-19 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
6MIP
 
 | |
6AXI
 
 | |
6BHX
 
 | | B. subtilis SsbA with DNA | | 分子名称: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Single-stranded DNA-binding protein A | | 著者 | Dubiel, K.D, Myers, A.R, Satyshur, K.A, Keck, J.L. | | 登録日 | 2017-10-31 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.936 Å) | | 主引用文献 | Structural Mechanisms of Cooperative DNA Binding by Bacterial Single-Stranded DNA-Binding Proteins.
J. Mol. Biol., 431, 2019
|
|
6MIL
 
 | | Crystal structure of AF9 YEATS domain in complex with histone H3K9bu | | 分子名称: | DI(HYDROXYETHYL)ETHER, Histone H3K9bu, MALONATE ION, ... | | 著者 | Vann, K.R, Klein, B.J, Kutateladze, T.G. | | 登録日 | 2018-09-19 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
8H69
 
 | | Cryo-EM structure of influenza RNA polymerase | | 分子名称: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*UP*AP*AP*AP*CP*UP*CP*CP*UP*GP*CP*UP*UP*UP*UP*GP*CP*U)-3'), ... | | 著者 | Li, H, Wu, Y, Liang, H, Liu, Y. | | 登録日 | 2022-10-16 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | An intermediate state allows influenza polymerase to switch smoothly between transcription and replication cycles.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6ING
 
 | |
5ZFZ
 
 | | Crystal structure of human DUX4 homeodomains bound to A12T DNA mutant | | 分子名称: | DNA (5'-D(*CP*CP*AP*CP*TP*AP*AP*CP*CP*TP*AP*TP*TP*CP*AP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*TP*GP*AP*AP*TP*AP*GP*GP*TP*TP*AP*GP*TP*GP*G)-3'), Double homeobox protein 4-like protein 4 | | 著者 | Li, Y.Y, Wu, B.X, Gan, J.H. | | 登録日 | 2018-03-07 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for multiple gene regulation by human DUX4.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
6BHW
 
 | | B. subtilis SsbA | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Single-stranded DNA-binding protein A | | 著者 | Dubiel, K.D, Myers, A.R, Satyshur, K.A, Keck, J.L. | | 登録日 | 2017-10-31 | | 公開日 | 2018-12-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.208 Å) | | 主引用文献 | Structural Mechanisms of Cooperative DNA Binding by Bacterial Single-Stranded DNA-Binding Proteins.
J. Mol. Biol., 431, 2019
|
|
4DUS
 
 | | Structure of Bace-1 (Beta-Secretase) in complex with N-((2S,3R)-1-(4-fluorophenyl)-3-hydroxy-4-((6'-neopentyl-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridin]-4'-yl)amino)butan-2-yl)acetamide | | 分子名称: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | 著者 | Sickmier, E.A. | | 登録日 | 2012-02-22 | | 公開日 | 2012-10-10 | | 最終更新日 | 2014-07-02 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A Potent and Orally Efficacious, Hydroxyethylamine-Based Inhibitor of beta-Secretase.
ACS Med Chem Lett, 3, 2012
|
|
4E93
 
 | | Crystal structure of human Feline Sarcoma Viral Oncogene Homologue (v-FES)in complex with TAE684 | | 分子名称: | 5-CHLORO-N-[2-METHOXY-4-[4-(4-METHYLPIPERAZIN-1-YL)PIPERIDIN-1-YL]PHENYL]-N'-(2-PROPAN-2-YLSULFONYLPHENYL)PYRIMIDINE-2,4-DIAMINE, Tyrosine-protein kinase Fes/Fps | | 著者 | Filippakopoulos, P, Salah, E, Miduturu, C.V, Fedorov, O, Cooper, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2012-03-20 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Small-Molecule Inhibitors of the c-Fes Protein-Tyrosine Kinase.
Chem.Biol., 19, 2012
|
|
4LY9
 
 | | Human GKRP complexed to AMG-1694 [(2R)-1,1,1-trifluoro-2-{4-[(2S)-2-{[(3S)-3-methylmorpholin-4-yl]methyl}-4-(thiophen-2-ylsulfonyl)piperazin-1-yl]phenyl}propan-2-ol] and sorbitol-6-phosphate | | 分子名称: | (2R)-1,1,1-trifluoro-2-{4-[(2S)-2-{[(3S)-3-methylmorpholin-4-yl]methyl}-4-(thiophen-2-ylsulfonyl)piperazin-1-yl]phenyl}propan-2-ol, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | 著者 | Jordan, S.R. | | 登録日 | 2013-07-30 | | 公開日 | 2013-11-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Antidiabetic effects of glucokinase regulatory protein small-molecule disruptors.
Nature, 504, 2013
|
|
5ZFY
 
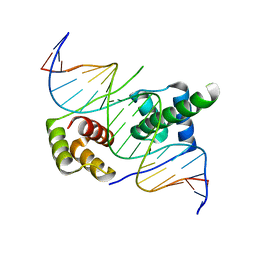 | | Crystal structure of human DUX4 homeodomains bound to A12C DNA mutant | | 分子名称: | DNA (5'-D(*CP*CP*AP*CP*TP*AP*AP*CP*CP*TP*AP*CP*TP*CP*AP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*TP*GP*AP*GP*TP*AP*GP*GP*TP*TP*AP*GP*TP*GP*G)-3'), Double homeobox protein 4-like protein 4 | | 著者 | Li, Y.Y, Wu, B.X, Gan, J.H. | | 登録日 | 2018-03-07 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for multiple gene regulation by human DUX4.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
5ZFW
 
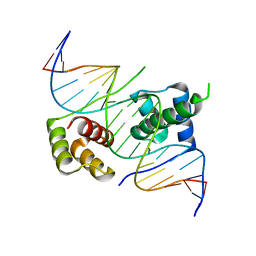 | | Crystal structure of human DUX4 homeodomains bound to A11G DNA mutant | | 分子名称: | DNA (5'-D(*CP*CP*AP*CP*TP*AP*AP*CP*CP*TP*GP*AP*TP*CP*AP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*TP*GP*AP*TP*CP*AP*GP*GP*TP*TP*AP*GP*TP*GP*G)-3'), Double homeobox protein 4-like protein 4 | | 著者 | Li, Y.Y, Wu, B.X, Gan, J.H. | | 登録日 | 2018-03-07 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.103 Å) | | 主引用文献 | Structural basis for multiple gene regulation by human DUX4.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
8HBT
 
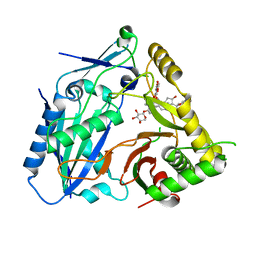 | | AmAT7-3 mutant A310G | | 分子名称: | AmAT7-3-A310G, Astragaloside IV | | 著者 | Wang, L.L. | | 登録日 | 2022-10-31 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Characterization and structure-based protein engineering of a regiospecific saponin acetyltransferase from Astragalus membranaceus.
Nat Commun, 14, 2023
|
|
8H8I
 
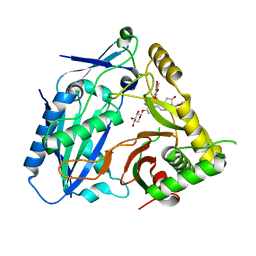 | | Triterpenoid saponin acetyltransferase, AmAT7-3 | | 分子名称: | AmAT7-3, Astragaloside IV | | 著者 | Wang, L.L. | | 登録日 | 2022-10-22 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Characterization and structure-based protein engineering of a regiospecific saponin acetyltransferase from Astragalus membranaceus.
Nat Commun, 14, 2023
|
|
4GNF
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3 peptide 1-15 | | 分子名称: | Histone H3.3, Histone-lysine N-methyltransferase NSD3, ZINC ION | | 著者 | Li, F, He, C, Wu, J, Shi, Y. | | 登録日 | 2012-08-17 | | 公開日 | 2013-01-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
4GNG
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3K9me3 peptide | | 分子名称: | GLYCEROL, Histone H3.3, Histone-lysine N-methyltransferase NSD3, ... | | 著者 | Li, F, He, C, Wu, J, Shi, Y. | | 登録日 | 2012-08-17 | | 公開日 | 2013-01-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
3TO4
 
 | | Structure of mouse Valpha14Vbeta2-mouseCD1d-alpha-Galactosylceramide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | 著者 | Patel, O, Rossjohn, J. | | 登録日 | 2011-09-04 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Vbeta2 natural killer T cell antigen receptor-mediated recognition of CD1d-glycolipid antigen.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8HMH
 
 | | The closed state of RGLG2-VWA | | 分子名称: | E3 ubiquitin-protein ligase RGLG2, MAGNESIUM ION | | 著者 | Wang, Q. | | 登録日 | 2022-12-03 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | The regulation of RGLG2-VWA by Ca 2+ ions.
Biochim Biophys Acta Proteins Proteom, 1872, 2024
|
|
