6PET
 
 | | Crystal structure of 8-hydroxychromene compound 30 bound to estrogen receptor alpha | | 分子名称: | (2S)-2-(4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-3-(3-hydroxyphenyl)-4-methyl-2H-1-benzopyran-8-ol, (2S)-3-(3-hydroxyphenyl)-2-(4-iodophenyl)-4-methyl-2H-1-benzopyran-6-ol, CHLORIDE ION, ... | | 著者 | Kiefer, J.R, Vinogradova, M, Liang, J, Wang, X, Zbieg, J, Labadie, S.S, Zhang, B, Li, J, Liang, W. | | 登録日 | 2019-06-20 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.203 Å) | | 主引用文献 | Discovery of a C-8 hydroxychromene as a potent degrader of estrogen receptor alpha with improved rat oral exposure over GDC-0927.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6PFM
 
 | | Crystal structure of GDC-0927 bound to estrogen receptor alpha | | 分子名称: | (2S)-2-(4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-3-(3-hydroxyphenyl)-4-methyl-2H-1-benzopyran-6-ol, Estrogen receptor | | 著者 | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Wang, X, Zbieg, J.R, Labadie, S.S, Li, J, Ray, N.C, Ortwine, D. | | 登録日 | 2019-06-21 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Discovery of a C-8 hydroxychromene as a potent degrader of estrogen receptor alpha with improved rat oral exposure over GDC-0927.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
7BUY
 
 | | The crystal structure of COVID-19 main protease in complex with carmofur | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, hexylcarbamic acid | | 著者 | Zhao, Y, Zhang, B, Jin, Z, Liu, X, Yang, H, Rao, Z. | | 登録日 | 2020-04-08 | | 公開日 | 2020-04-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7BQY
 
 | | THE CRYSTAL STRUCTURE OF COVID-19 MAIN PROTEASE IN COMPLEX WITH AN INHIBITOR N3 at 1.7 angstrom | | 分子名称: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | 著者 | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | 登録日 | 2020-03-26 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
5MK5
 
 | | Structures of DHBN domain of human BLM helicase | | 分子名称: | Bloom syndrome protein, IODIDE ION, POTASSIUM ION | | 著者 | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | 登録日 | 2016-12-02 | | 公開日 | 2017-03-01 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
5WTE
 
 | | Cryo-EM structure for Hepatitis A virus full particle | | 分子名称: | VP1, VP2, VP3 | | 著者 | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | 登録日 | 2016-12-11 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WTG
 
 | | Crystal structure of the Fab fragment of anti-HAV antibody R10 | | 分子名称: | FAB Heavy chain, FAB Light chain | | 著者 | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | 登録日 | 2016-12-11 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.907 Å) | | 主引用文献 | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XMY
 
 | |
5Y20
 
 | |
5YC4
 
 | |
6XVH
 
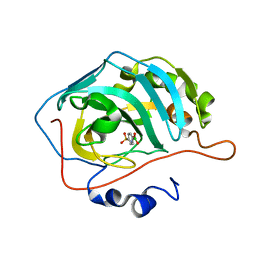 | | carbonic anhydrase with bound arylsulfonamide- boronic acid | | 分子名称: | (4-sulfamoylphenyl)boronic acid, Carbonic anhydrase 2, ZINC ION | | 著者 | Perez, C, Zhang, B. | | 登録日 | 2020-01-22 | | 公開日 | 2021-02-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | An easy to assemble, fully modulable, selectiv and
colorimetric artificial bio-sensor for fingerprintting
the recognition of neurotransmitters
To Be Published
|
|
5YC3
 
 | |
6WOK
 
 | | Crystal structure of estrogen receptor alpha in complex with receptor degrader 6 | | 分子名称: | (1R,3R)-1-(2,6-difluoro-4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-2-(2-fluoro-2-methylpropyl)-3-methyl-2,3,4,9-tetrahydro-1H-beta-carboline, (2S)-3-(3-hydroxyphenyl)-2-(4-iodophenyl)-4-methyl-2H-1-benzopyran-6-ol, Estrogen receptor | | 著者 | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Wang, X, Labadie, S. | | 登録日 | 2020-04-24 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.309 Å) | | 主引用文献 | Discovery of GNE-149 as a Full Antagonist and Efficient Degrader of Estrogen Receptor alpha for ER+ Breast Cancer.
Acs Med.Chem.Lett., 11, 2020
|
|
6CB7
 
 | |
6CB6
 
 | |
6LU7
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor N3 | | 分子名称: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | 著者 | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | 登録日 | 2020-01-26 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
4S1R
 
 | | Crystal structure of a VRC01-lineage antibody, 45-VRC01.H08.F-117225, in complex with clade A/E HIV-1 gp120 core | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab of VRC01 light chain, Fab of VRC01-lineage antibody,45-VRC01.H08.F-117225 heavy chain, ... | | 著者 | Kwon, Y.D, Yang, Y, Zhang, B, Kwong, P.D. | | 登録日 | 2015-01-14 | | 公開日 | 2015-04-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.214 Å) | | 主引用文献 | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell(Cambridge,Mass.), 161, 2015
|
|
4S1Q
 
 | | Crystal structure of a VRC01-lineage antibody, 45-VRC01.H03+06.D-001739, in complex with clade A/E HIV-1 gp120 core | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab of VRC01 light chain, Fab of VRC01-lineage antibody,45-VRC01.H03+06.D-001739 heavy chain, ... | | 著者 | Kwon, Y.D, Yang, Y, Zhang, B, Kwong, P.D. | | 登録日 | 2015-01-14 | | 公開日 | 2015-04-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell(Cambridge,Mass.), 161, 2015
|
|
4S1S
 
 | | Crystal structure of a VRC01-lineage antibody, 45-VRC01.H5.F-185917, in complex with clade A/E HIV-1 gp120 core | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fab of VRC01 light chain, ... | | 著者 | Kwon, Y.D, Yang, Y, Zhang, B, Kwong, P.D. | | 登録日 | 2015-01-14 | | 公開日 | 2015-04-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.39 Å) | | 主引用文献 | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell(Cambridge,Mass.), 161, 2015
|
|
4KR0
 
 | | Complex structure of MERS-CoV spike RBD bound to CD26 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | 著者 | Lu, G, Hu, Y, Wang, Q, Qi, J, Gao, F, Li, Y, Zhang, Y, Zhang, W, Yuan, Y, Zhang, B, Shi, Y, Yan, J, Gao, G.F. | | 登録日 | 2013-05-15 | | 公開日 | 2013-07-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.702 Å) | | 主引用文献 | Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26.
Nature, 500, 2013
|
|
4KQZ
 
 | | structure of the receptor binding domain (RBD) of MERS-CoV spike | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | 著者 | Lu, G, Hu, Y, Wang, Q, Qi, J, Gao, F, Li, Y, Zhang, Y, Zhang, W, Yuan, Y, Bao, J, Zhang, B, Shi, Y, Yan, J, Gao, G.F. | | 登録日 | 2013-05-15 | | 公開日 | 2013-07-10 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.514 Å) | | 主引用文献 | Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26.
Nature, 500, 2013
|
|
5K6F
 
 | | Crystal structure of prefusion-stabilized RSV F single-chain 9-19 DS-Cav1 variant. | | 分子名称: | Fusion glycoprotein F0 | | 著者 | Joyce, M.G, Zhang, B, Lai, Y.T, Mascola, J.R, Kwong, P.D. | | 登録日 | 2016-05-24 | | 公開日 | 2016-08-10 | | 最終更新日 | 2016-09-21 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5K6C
 
 | | Crystal structure of prefusion-stabilized RSV F single-chain 9-10 DS-Cav1 variant. | | 分子名称: | Fusion glycoprotein F0,Fusion glycoprotein F0, SULFATE ION | | 著者 | Joyce, M.G, Zhang, B, Lai, Y.T, Mascola, J.R, Kwong, P.D. | | 登録日 | 2016-05-24 | | 公開日 | 2016-10-12 | | 最終更新日 | 2016-10-19 | | 実験手法 | X-RAY DIFFRACTION (3.576 Å) | | 主引用文献 | Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV.
Nat.Struct.Mol.Biol., 23, 2016
|
|
8GTY
 
 | |
5K6H
 
 | |
