7Y7J
 
 | | SARS-CoV-2 S trimer in complex with 1F Fab | | 分子名称: | 1F VH, 1F VL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhao, S, Liu, F, Yang, X, Zhong, G. | | 登録日 | 2022-06-22 | | 公開日 | 2023-02-22 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | A core epitope targeting antibody of SARS-CoV-2.
Protein Cell, 14, 2023
|
|
7Y7K
 
 | | SARS-CoV-2 RBD in complex with 1F Fab | | 分子名称: | 1F VH, 1F VL, Spike protein S1 | | 著者 | Zhao, S, Liu, F, Yang, X, Zhong, G. | | 登録日 | 2022-06-22 | | 公開日 | 2023-02-22 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | A core epitope targeting antibody of SARS-CoV-2.
Protein Cell, 14, 2023
|
|
8HPM
 
 | | LpqY-SugABC in state 2 | | 分子名称: | ABC sugar transporter, permease component, ABC transporter, ... | | 著者 | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | 登録日 | 2022-12-12 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3.82 Å) | | 主引用文献 | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPN
 
 | | LpqY-SugABC in state 3 | | 分子名称: | ABC sugar transporter, permease component, ABC transporter, ... | | 著者 | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | 登録日 | 2022-12-12 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (4.55 Å) | | 主引用文献 | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPS
 
 | | LpqY-SugABC in state 5 | | 分子名称: | ABC sugar transporter, permease component, ABC transporter, ... | | 著者 | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | 登録日 | 2022-12-12 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3.51 Å) | | 主引用文献 | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPL
 
 | | LpqY-SugABC in state 1 | | 分子名称: | ABC sugar transporter, permease component, ABC transporter, ... | | 著者 | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | 登録日 | 2022-12-12 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (4.29 Å) | | 主引用文献 | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPR
 
 | | LpqY-SugABC in state 4 | | 分子名称: | ABC sugar transporter, permease component, ABC transporter, ... | | 著者 | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | 登録日 | 2022-12-12 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8JRE
 
 | | Cryo-EM structure of a designed AAV8-based vector | | 分子名称: | Capsid protein | | 著者 | Ke, X, Luo, S, Zheng, Q, Jiang, H, Liu, F, Sun, X. | | 登録日 | 2023-06-16 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (2.08 Å) | | 主引用文献 | An adeno-associated virus variant enabling efficient ocular-directed gene delivery across species.
Nat Commun, 15, 2024
|
|
7YVE
 
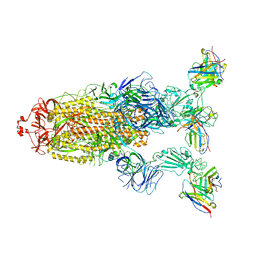 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH027 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH027 Fab heavy chain, ... | | 著者 | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | 登録日 | 2022-08-19 | | 公開日 | 2023-06-28 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVP
 
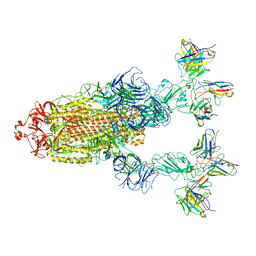 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH272/281 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH272 Fab heavy chain, ... | | 著者 | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | 登録日 | 2022-08-19 | | 公開日 | 2023-06-28 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVG
 
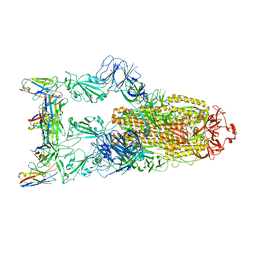 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH132 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH132 Fab heavy chain, ... | | 著者 | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | 登録日 | 2022-08-19 | | 公開日 | 2023-06-28 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVK
 
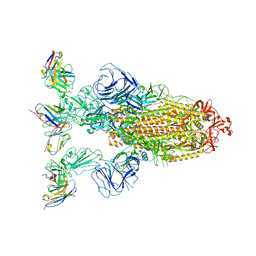 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH272 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH272 Fab heavy chain, ... | | 著者 | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | 登録日 | 2022-08-19 | | 公開日 | 2023-06-28 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVO
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH027/132 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH132 Fab heavy chain, ... | | 著者 | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | 登録日 | 2022-08-19 | | 公開日 | 2023-06-28 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVN
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH281 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH281 Fab heavy chain, ... | | 著者 | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | 登録日 | 2022-08-19 | | 公開日 | 2023-06-28 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVH
 
 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH132 Fab | | 分子名称: | Spike glycoprotein, TH132 Fab heavy chain, TH132 Fab light chain | | 著者 | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | 登録日 | 2022-08-19 | | 公開日 | 2023-06-28 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVL
 
 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH272 Fab | | 分子名称: | Spike glycoprotein, TH272 Fab heavy chain, TH272 Fab light chain | | 著者 | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | 登録日 | 2022-08-19 | | 公開日 | 2023-06-28 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVM
 
 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH272 Fab | | 分子名称: | Spike glycoprotein, TH281 Fab heavy chain, TH281 Fab light chain | | 著者 | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | 登録日 | 2022-08-19 | | 公開日 | 2023-06-28 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVF
 
 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH027 Fab | | 分子名称: | Spike glycoprotein, TH027 Fab heavy chain, TH027 Fab light chain | | 著者 | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | 登録日 | 2022-08-19 | | 公開日 | 2023-06-28 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVI
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH236 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH236 Fab heavy chain, ... | | 著者 | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | 登録日 | 2022-08-19 | | 公開日 | 2023-06-28 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVJ
 
 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH236 Fab | | 分子名称: | Spike glycoprotein, TH236 Fab heavy chain, TH236 Fab light chain | | 著者 | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | 登録日 | 2022-08-19 | | 公開日 | 2023-06-28 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
5W81
 
 | | Phosphorylated, ATP-bound structure of zebrafish cystic fibrosis transmembrane conductance regulator (CFTR) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | 著者 | Zhang, Z, Liu, F, Chen, J. | | 登録日 | 2017-06-21 | | 公開日 | 2017-07-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Conformational Changes of CFTR upon Phosphorylation and ATP Binding.
Cell, 170, 2017
|
|
2ELA
 
 | | Crystal Structure of the PTB domain of human APPL1 | | 分子名称: | Adapter protein containing PH domain, PTB domain and leucine zipper motif 1 | | 著者 | Li, J, Mao, X, Dong, L.Q, Liu, F, Tong, L. | | 登録日 | 2007-03-27 | | 公開日 | 2007-05-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structures of the BAR-PH and PTB Domains of Human APPL1
Structure, 15, 2007
|
|
2ELB
 
 | | Crystal Structure of the BAR-PH domain of human APPL1 | | 分子名称: | Adapter protein containing PH domain, PTB domain and leucine zipper motif 1 | | 著者 | Li, J, Mao, X, Dong, L.Q, Liu, F, Tong, L. | | 登録日 | 2007-03-27 | | 公開日 | 2007-05-29 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structures of the BAR-PH and PTB Domains of Human APPL1
Structure, 15, 2007
|
|
2HS2
 
 | | Crystal structure of M46L mutant of HIV-1 protease complexed with TMC114 (darunavir) | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Weber, I.T, Kovalevsky, A.Y, Liu, F. | | 登録日 | 2006-07-20 | | 公開日 | 2006-10-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Ultra-high Resolution Crystal Structure of HIV-1 Protease Mutant Reveals Two Binding Sites for Clinical Inhibitor TMC114.
J.Mol.Biol., 363, 2006
|
|
8GS8
 
 | | cryo-EM structure of the human respiratory complex II | | 分子名称: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | 著者 | Du, Z, Zhou, X, Lai, Y, Xu, J, Zhang, Y, Zhou, S, Liu, F, Gao, Y, Gong, H, Rao, Z. | | 登録日 | 2022-09-05 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Structure of the human respiratory complex II.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
