4EUH
 
 | | Crystal structure of Clostridium acetobutulicum trans-2-enoyl-CoA reductase apo form | | 分子名称: | Putative reductase CA_C0462, SODIUM ION | | 著者 | Hu, K, Zhao, M, Zhang, T, Yang, S, Ding, J. | | 登録日 | 2012-04-25 | | 公開日 | 2012-11-28 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structures of trans-2-enoyl-CoA reductases from Clostridium acetobutylicum and Treponema denticola: insights into the substrate specificity and the catalytic mechanism
Biochem.J., 449, 2013
|
|
5H4B
 
 | | Crystal structure of Cbln4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-4 | | 著者 | Zhong, C, Shen, J, Zhang, H, Ding, J. | | 登録日 | 2016-10-31 | | 公開日 | 2017-09-13 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
8GTN
 
 | |
8GTK
 
 | |
8GTJ
 
 | |
3IWP
 
 | |
5WTT
 
 | | Structure of the 093G9 Fab in complex with the epitope peptide | | 分子名称: | Epitope peptide of Cyr61, Heavy chain of 093G9 Fab, Light chain of 093G9 Fab | | 著者 | Zhong, C, Hu, K, Shen, J, Ding, J. | | 登録日 | 2016-12-14 | | 公開日 | 2017-12-20 | | 最終更新日 | 2019-01-02 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Molecular basis for the recognition of CCN1 by monoclonal antibody 093G9.
J. Mol. Recognit., 30, 2017
|
|
3NFP
 
 | | Crystal structure of the Fab fragment of therapeutic antibody daclizumab in complex with IL-2Ra (CD25) ectodomain | | 分子名称: | Heavy chain of Fab fragment of daclizumab, Interleukin-2 receptor subunit alpha, Light chain of Fab fragment of daclizumab | | 著者 | Yang, H, Wang, J, Du, J, Zhong, C, Guo, Y, Ding, J. | | 登録日 | 2010-06-10 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Structural basis of immunosuppression by the therapeutic antibody daclizumab
Cell Res., 20, 2010
|
|
4HK6
 
 | |
4HK7
 
 | |
4HK5
 
 | | Crystal structure of Cordyceps militaris IDCase in apo form | | 分子名称: | Uracil-5-carboxylate decarboxylase, ZINC ION | | 著者 | Xu, S, Zhu, J, Ding, J. | | 登録日 | 2012-10-15 | | 公開日 | 2013-09-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4HJW
 
 | |
3NFS
 
 | | Crystal structure the Fab fragment of therapeutic antibody daclizumab | | 分子名称: | Heavy chain of Fab fragment of daclizumab, Light chain of Fab fragment of daclizumab | | 著者 | Yang, H, Wang, J, Du, J, Zhong, C, Guo, Y, Ding, J. | | 登録日 | 2010-06-10 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis of immunosuppression by the therapeutic antibody daclizumab
Cell Res., 20, 2010
|
|
4FG7
 
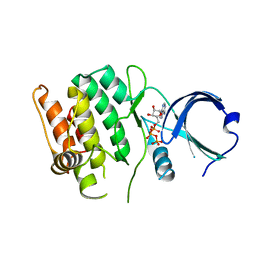 | | Crystal structure of human calcium/calmodulin-dependent protein kinase I 1-293 in complex with ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Calcium/calmodulin-dependent protein kinase type 1 | | 著者 | Zha, M, Zhong, C, Ou, Y, Wang, J, Han, L, Ding, J. | | 登録日 | 2012-06-04 | | 公開日 | 2013-01-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structures of human CaMKIalpha reveal insights into the regulation mechanism of CaMKI.
Plos One, 7, 2012
|
|
1QK6
 
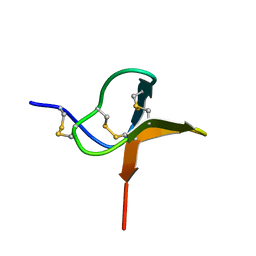 | | Solution structure of huwentoxin-I by NMR | | 分子名称: | HUWENTOXIN-I | | 著者 | Qu, Y, Liang, S, Ding, J, Liu, X, Zhang, R, Gu, X. | | 登録日 | 1999-07-10 | | 公開日 | 1999-08-20 | | 最終更新日 | 2019-01-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Proton Nuclear Magnetic Resonance Studies on Huwentoxin-I from the Venom of the Spider Selenocosmia Huwena:2.Three-Dimensional Structure in Solution
J.Protein Chem., 16, 1997
|
|
3V9R
 
 | | Crystal structure of Saccharomyces cerevisiae MHF complex | | 分子名称: | SULFATE ION, Uncharacterized protein YDL160C-A, Uncharacterized protein YOL086W-A | | 著者 | Yang, H, Zhang, T, Zhong, C, Li, H, Zhou, J, Ding, J. | | 登録日 | 2011-12-28 | | 公開日 | 2012-02-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Saccharomyces Cerevisiae MHF Complex Structurally Resembles the Histones (H3-H4)(2) Heterotetramer and Functions as a Heterotetramer
Structure, 20, 2012
|
|
4HKA
 
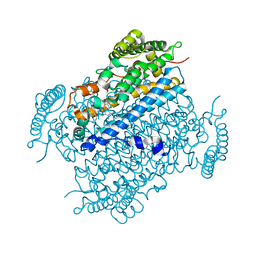 | |
1L3K
 
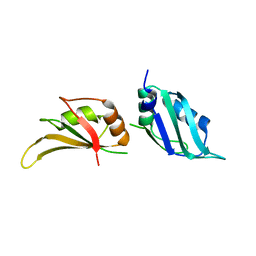 | | UP1, THE TWO RNA-RECOGNITION MOTIF DOMAIN OF HNRNP A1 | | 分子名称: | HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN A1 | | 著者 | Vitali, J, Ding, J, Jiang, J, Zhang, Y, Krainer, A.R, Xu, R.-M. | | 登録日 | 2002-02-27 | | 公開日 | 2002-04-17 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Correlated alternative side chain conformations in the RNA-recognition motif of heterogeneous nuclear ribonucleoprotein A1.
Nucleic Acids Res., 30, 2002
|
|
1DLO
 
 | | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | 分子名称: | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 REVERSE TRANSCRIPTASE | | 著者 | Hsiou, Y, Ding, J, Das, K, Hughes, S, Arnold, E. | | 登録日 | 1996-04-17 | | 公開日 | 1996-08-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of unliganded HIV-1 reverse transcriptase at 2.7 A resolution: implications of conformational changes for polymerization and inhibition mechanisms.
Structure, 4, 1996
|
|
3OPT
 
 | | Crystal structure of the Rph1 catalytic core with a-ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, DNA damage-responsive transcriptional repressor RPH1, NICKEL (II) ION | | 著者 | Chang, Y, Wu, J, Tong, X, Zhou, J, Ding, J. | | 登録日 | 2010-09-02 | | 公開日 | 2010-12-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of the catalytic core of Saccharomyces cerevesiae histone demethylase Rph1: insights into the substrate specificity and catalytic mechanism
Biochem.J., 433, 2011
|
|
3OPW
 
 | | Crystal Structure of the Rph1 catalytic core | | 分子名称: | DNA damage-responsive transcriptional repressor RPH1 | | 著者 | Chang, Y, Wu, J, Tong, X, Zhou, J, Ding, J. | | 登録日 | 2010-09-02 | | 公開日 | 2010-12-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the catalytic core of Saccharomyces cerevesiae histone demethylase Rph1: insights into the substrate specificity and catalytic mechanism
Biochem.J., 433, 2011
|
|
6LPU
 
 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with L-2-hydroxyglutarate (L-2-HG) | | 分子名称: | (2S)-2-HYDROXYPENTANEDIOIC ACID, D-2-hydroxyglutarate dehydrogenase, mitochondrial, ... | | 著者 | Yang, J, Zhu, H, Ding, J. | | 登録日 | 2020-01-12 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.923 Å) | | 主引用文献 | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPQ
 
 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with D-malate (D-MAL) | | 分子名称: | D-2-hydroxyglutarate dehydrogenase, mitochondrial, D-MALATE, ... | | 著者 | Yang, J, Zhu, H, Ding, J. | | 登録日 | 2020-01-12 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPN
 
 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in apo form | | 分子名称: | D-2-hydroxyglutarate dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Yang, J, Zhu, H, Ding, J. | | 登録日 | 2020-01-12 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.206 Å) | | 主引用文献 | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
1XTR
 
 | |
