6HP9
 
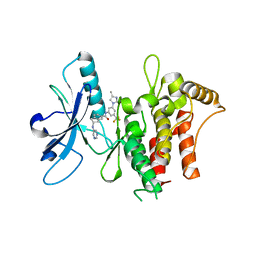 | | Structure of the kinase domain of human DDR1 in complex with a 2-Amino-2,3-Dihydro-1H-Indene-5-Carboxamide-based inhibitor | | 分子名称: | (2~{R})-~{N}-[3-(4-methylimidazol-1-yl)-5-(trifluoromethyl)phenyl]-2-(pyrimidin-5-ylamino)-2,3-dihydro-1~{H}-indene-5-carboxamide, Epithelial discoidin domain-containing receptor 1 | | 著者 | Pinkas, D.M, Fox, A.E, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N, Structural Genomics Consortium (SGC) | | 登録日 | 2018-09-19 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | 2-Amino-2,3-dihydro-1H-indene-5-carboxamide-Based Discoidin Domain Receptor 1 (DDR1) Inhibitors: Design, Synthesis, and in Vivo Antipancreatic Cancer Efficacy.
J.Med.Chem., 62, 2019
|
|
8X3R
 
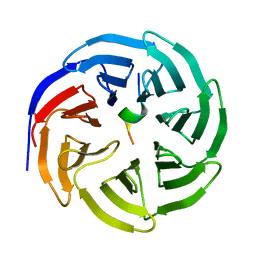 | |
8X3S
 
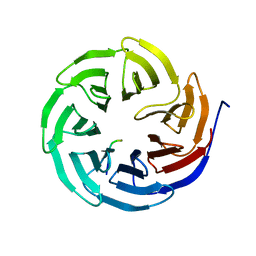 | | Crystal structure of human WDR5 in complex with PTEN | | 分子名称: | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, WD repeat-containing protein 5 | | 著者 | Liu, Y, Huang, X, Shang, X. | | 登録日 | 2023-11-14 | | 公開日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | The NTE domain of PTEN alpha / beta promotes cancer progression by interacting with WDR5 via its SSSRRSS motif.
Cell Death Dis, 15, 2024
|
|
5YAZ
 
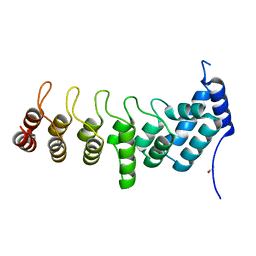 | | Crystal structure of the ANKRD domain of KANK1 | | 分子名称: | ACETATE ION, KN motif and ankyrin repeat domains 1 | | 著者 | Wei, Z, Pan, W. | | 登録日 | 2017-09-02 | | 公開日 | 2017-12-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural insights into ankyrin repeat-mediated recognition of the kinesin motor protein KIF21A by KANK1, a scaffold protein in focal adhesion.
J. Biol. Chem., 293, 2018
|
|
5YAY
 
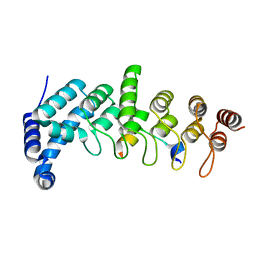 | | Crystal structure of KANK1/KIF21A complex | | 分子名称: | KN motif and ankyrin repeat domains 1, Kinesin-like protein KIF21A | | 著者 | Wei, Z, Pan, W. | | 登録日 | 2017-09-02 | | 公開日 | 2017-12-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural insights into ankyrin repeat-mediated recognition of the kinesin motor protein KIF21A by KANK1, a scaffold protein in focal adhesion.
J. Biol. Chem., 293, 2018
|
|
7YKC
 
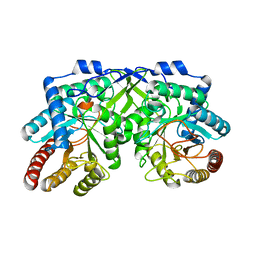 | |
8JLX
 
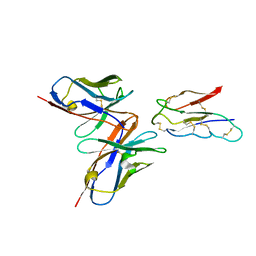 | | CCHFV envelope protein Gc in complex with Gc13 | | 分子名称: | Glycoprotein C,CCHFV Gc fusion loops, Mouse antibody Gc13 heavy chain, Mouse antibody Gc13 light chain | | 著者 | Chong, T, Cao, S. | | 登録日 | 2023-06-03 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Neutralizing monoclonal antibodies against the Gc fusion loop region of Crimean-Congo hemorrhagic fever virus.
Plos Pathog., 20, 2024
|
|
8JLW
 
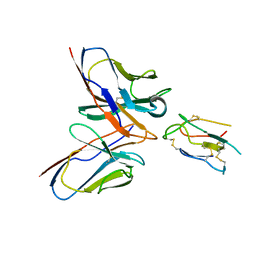 | | CCHFV envelope protein Gc in complex with Gc8 | | 分子名称: | Glycoprotein C,CCHFV envelope protein Gc fusion loops, Mouse antibody Gc8 heavy chain, Mouse antibody Gc8 light chain | | 著者 | Chong, T, Cao, S. | | 登録日 | 2023-06-03 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Neutralizing monoclonal antibodies against the Gc fusion loop region of Crimean-Congo hemorrhagic fever virus.
Plos Pathog., 20, 2024
|
|
8JKD
 
 | |
8JF4
 
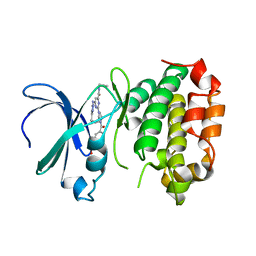 | | The crystal structure of human AURKA kinase domain in complex with AURKA-compound 9 | | 分子名称: | 2-[4-[4-[bis(oxidanylidene)-$l^5-sulfanyl]oxyphenyl]carbonylpiperazin-1-yl]-6-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]-N-prop-2-ynyl-pyrimidine-4-carboxamide, Aurora kinase A | | 著者 | Zhu, C.J. | | 登録日 | 2023-05-17 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.89288354 Å) | | 主引用文献 | Global Reactivity Profiling of the Catalytic Lysine in Human Kinome for Covalent Inhibitor Development.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8JF3
 
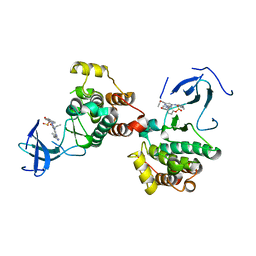 | | C-Src in complex with compound 9 | | 分子名称: | 2-[4-[4-[bis(oxidanylidene)-$l^5-sulfanyl]oxyphenyl]carbonylpiperazin-1-yl]-6-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]-N-prop-2-ynyl-pyrimidine-4-carboxamide, Proto-oncogene tyrosine-protein kinase Src | | 著者 | Zhang, Z.M, Huang, H.S. | | 登録日 | 2023-05-17 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.84647632 Å) | | 主引用文献 | Global Reactivity Profiling of the Catalytic Lysine in Human Kinome for Covalent Inhibitor Development.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8JG8
 
 | |
7C83
 
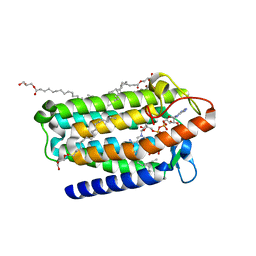 | | Crystal structure of an integral membrane steroid 5-alpha-reductase PbSRD5A | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-oxo-5-alpha-steroid 4-dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Ren, R.B, Han, Y.F, Xiao, Q.J, Deng, D. | | 登録日 | 2020-05-28 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of steroid reductase SRD5A reveals conserved steroid reduction mechanism.
Nat Commun, 12, 2021
|
|
7EFT
 
 | |
6IGZ
 
 | | Structure of PSI-LHCI | | 分子名称: | (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | 著者 | Xiong, P, Xiaochun, Q. | | 登録日 | 2018-09-27 | | 公開日 | 2019-02-13 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Structure of a green algal photosystem I in complex with a large number of light-harvesting complex I subunits.
Nat Plants, 5, 2019
|
|
7DL9
 
 | |
7DLA
 
 | |
7DWN
 
 | | Crystal structure of Vibrio fischeri DarR in complex with DNA reveals the transcriptional activation mechanism of LTTR family members | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Predicted DNA-binding transcriptional regulator | | 著者 | Wang, W.W, Wu, H, He, J.H, Yu, F. | | 登録日 | 2021-01-17 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Crystal structure details of Vibrio fischeri DarR and mutant DarR-M202I from LTTR family reveals their activation mechanism.
Int.J.Biol.Macromol., 183, 2021
|
|
7DWO
 
 | |
7EPW
 
 | |
7EPV
 
 | |
5Y93
 
 | | Crystal Structure Analysis of the BRD4 | | 分子名称: | 1,2-ETHANEDIOL, 2-[[5-[(5-bromanyl-2-methoxy-phenyl)sulfonylamino]-3-methyl-1,2-benzoxazol-6-yl]oxy]-N-(2-morpholin-4-ylethyl)ethanamide, Bromodomain-containing protein 4, ... | | 著者 | Xu, Y, Zhang, Y, Song, M, Wang, C. | | 登録日 | 2017-08-22 | | 公開日 | 2018-06-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y8W
 
 | | Crystal Structure Analysis of the BRD4 | | 分子名称: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-N-(3-methyl-6-oxidanyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | 著者 | Xu, Y, Zhang, Y, Song, M, Wang, C. | | 登録日 | 2017-08-21 | | 公開日 | 2018-06-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y94
 
 | | Crystal Structure Analysis of the BRD4 | | 分子名称: | 5-bromanyl-2-methoxy-N-[3-methyl-6-(methylamino)-1,2-benzoxazol-5-yl]benzenesulfonamide, Bromodomain-containing protein 4, GLYCEROL, ... | | 著者 | Xu, Y, Zhang, Y, Song, M, Wang, C. | | 登録日 | 2017-08-22 | | 公開日 | 2018-06-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5YQX
 
 | | Crystal Structure Analysis of the BRD4 | | 分子名称: | (2R)-2-(cyclopropylmethyl)-7-(3,5-dimethyl-1,2-oxazol-4-yl)-4H-1,4-benzoxazin-3-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | 著者 | Xue, X, Zhang, Y, Wang, C, Song, M. | | 登録日 | 2017-11-08 | | 公開日 | 2018-11-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Benzoxazinone-containing 3,5-dimethylisoxazole derivatives as BET bromodomain inhibitors for treatment of castration-resistant prostate cancer.
Eur.J.Med.Chem., 152, 2018
|
|
