9LGM
 
 | | Cryo-EM structure of GPR4 complexed with Gs in pH8.0 | | 分子名称: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Yue, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | 登録日 | 2025-01-10 | | 公開日 | 2025-04-23 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (2.84 Å) | | 主引用文献 | Structural basis of stepwise proton sensing-mediated GPCR activation.
Cell Res., 35, 2025
|
|
9JQR
 
 | |
9JQP
 
 | |
9JQ6
 
 | | Cryo-EM structure of BTN2A1 in complex with antagonist antibody TH002 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BTN2A1 antagonist antibody TH002-Fab heavy chain, ... | | 著者 | Zhang, M, Wang, Y.Q, Xiao, J.Y. | | 登録日 | 2024-09-27 | | 公開日 | 2025-06-18 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | Structures of butyrophilin multimers reveal a plier-like mechanism for V gamma 9V delta 2 T cell receptor activation.
Immunity, 2025
|
|
9JQQ
 
 | | Cryo-EM structure of the HMBPP-primed BTN3A1-BTN3A2-BTN2A1 complex | | 分子名称: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhang, M, Wang, Y.Q, Xiao, J.Y. | | 登録日 | 2024-09-28 | | 公開日 | 2025-06-18 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Structures of butyrophilin multimers reveal a plier-like mechanism for V gamma 9V delta 2 T cell receptor activation.
Immunity, 2025
|
|
4UT7
 
 | | CRYSTAL STRUCTURE OF THE SCFV FRAGMENT OF THE BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11 | | 分子名称: | BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11 | | 著者 | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | 登録日 | 2014-07-18 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
5XNM
 
 | | Structure of unstacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | 著者 | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | 登録日 | 2017-05-23 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
5XNL
 
 | | Structure of stacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | 著者 | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | 登録日 | 2017-05-23 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
5XNN
 
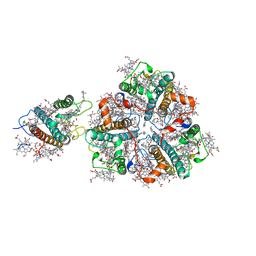 | | Structure of M-LHCII and CP24 complexes in the stacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | 著者 | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | 登録日 | 2017-05-23 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
7N5U
 
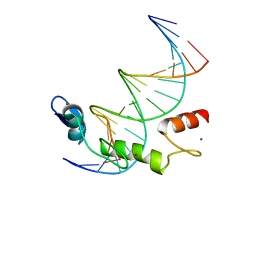 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 21) | | 分子名称: | DNA Strain II, DNA Strand I, ZINC ION, ... | | 著者 | Horton, J.R, Ren, R, Cheng, X. | | 登録日 | 2021-06-06 | | 公開日 | 2022-06-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
7N5W
 
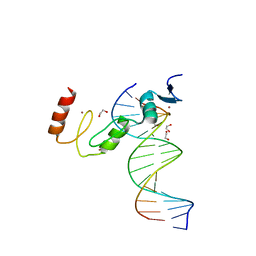 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 23) | | 分子名称: | 1,2-ETHANEDIOL, DNA Strand I, DNA Strand II, ... | | 著者 | Horton, J.R, Ren, R, Cheng, X. | | 登録日 | 2021-06-06 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
7N5V
 
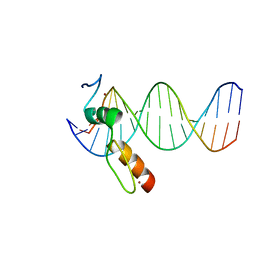 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 20) | | 分子名称: | DNA Strand I, DNA Strand II, ZINC ION, ... | | 著者 | Horton, J.R, Ren, R, Cheng, X. | | 登録日 | 2021-06-06 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.08 Å) | | 主引用文献 | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
7F6V
 
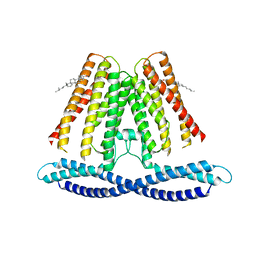 | | Cryo-EM structure of the human TACAN channel in a closed state | | 分子名称: | CHOLESTEROL, Ion channel TACAN | | 著者 | Chen, X.Z, Wang, Y.J, Li, Y, Yang, X, Shen, Y.Q. | | 登録日 | 2021-06-25 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | Cryo-EM structure of the human TACAN in a closed state.
Cell Rep, 38, 2022
|
|
4V40
 
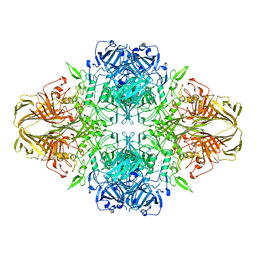 | | BETA-GALACTOSIDASE | | 分子名称: | BETA-GALACTOSIDASE, MAGNESIUM ION | | 著者 | Jacobson, R.H, Zhang, X, Dubose, R.F, Matthews, B.W. | | 登録日 | 1994-07-18 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Three-dimensional structure of beta-galactosidase from E. coli.
Nature, 369, 1994
|
|
8AG1
 
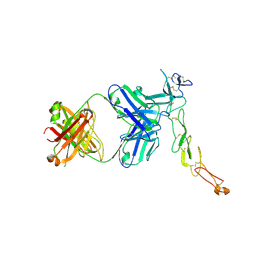 | | Crystal structure of a novel OX40 antibody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Novel OX40 antibody heavy chain, Novel OX40 antibody light chain, ... | | 著者 | Gao, H, Zhou, A. | | 登録日 | 2022-07-19 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3.303 Å) | | 主引用文献 | Structural Basis of a Novel Agonistic Anti-OX40 Antibody.
Biomolecules, 12, 2022
|
|
4UT9
 
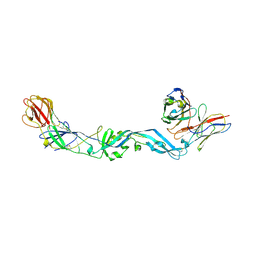 | | Crystal structure of dengue 2 virus envelope glycoprotein dimer in complex with the ScFv fragment of the broadly neutralizing human antibody EDE1 C10 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C10, ENVELOPE GLYCOPROTEIN E | | 著者 | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | 登録日 | 2014-07-18 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
4UTC
 
 | | Crystal structure of dengue 2 virus envelope glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN E, ... | | 著者 | Kikuti, C, Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | 登録日 | 2014-07-18 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (3.08 Å) | | 主引用文献 | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
4UTB
 
 | | Crystal structure of dengue 2 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE2 A11 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11, ... | | 著者 | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | 登録日 | 2014-07-18 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.85 Å) | | 主引用文献 | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
6LNK
 
 | | Candida albicans Fructose-1,6-bisphosphate aldolase | | 分子名称: | 1,2-ETHANEDIOL, Fructose-bisphosphate aldolase, ZINC ION | | 著者 | Huang, Y, Cao, H, Ren, Y, Wan, J. | | 登録日 | 2019-12-30 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.639 Å) | | 主引用文献 | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
9LDS
 
 | |
9ODR
 
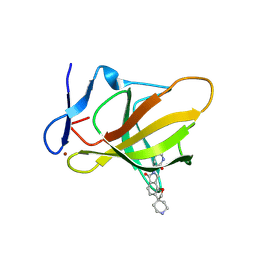 | | Structure of CRBN TBD bound to compound C1 | | 分子名称: | (3S)-3-(6-oxo-6,8-dihydro-2H,7H-spiro[furo[2,3-e]isoindole-3,4'-piperidin]-7-yl)piperidine-2,6-dione, Protein cereblon, ZINC ION | | 著者 | Strickland, C, Rice, C. | | 登録日 | 2025-04-27 | | 公開日 | 2025-05-28 | | 最終更新日 | 2025-06-25 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Discovery and Characterization of PVTX-321 as a Potent and Orally Bioavailable Estrogen Receptor Degrader for ER+/HER2- Breast Cancer.
J.Med.Chem., 68, 2025
|
|
9ODS
 
 | | Structure of CRBN TBD bound to compound C3 | | 分子名称: | (1P)-1-[(4S)-8H-spiro[furo[2,3-c]imidazo[1,2-a]pyridine-7,4'-piperidin]-3-yl]pyrimidine-2,4(1H,3H)-dione, Protein cereblon, ZINC ION | | 著者 | Strickland, C, Rice, C. | | 登録日 | 2025-04-27 | | 公開日 | 2025-05-28 | | 最終更新日 | 2025-06-25 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Discovery and Characterization of PVTX-321 as a Potent and Orally Bioavailable Estrogen Receptor Degrader for ER+/HER2- Breast Cancer.
J.Med.Chem., 68, 2025
|
|
6LEB
 
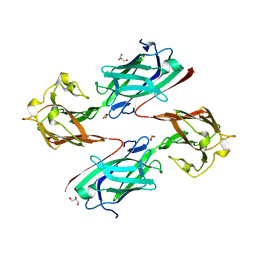 | | Staphylococcus aureus surface protein SdrC mutant-P366H | | 分子名称: | GLYCEROL, MAGNESIUM ION, Ser-Asp rich fibrinogen-binding, ... | | 著者 | Hang, T, Zhang, M, Wang, J. | | 登録日 | 2019-11-25 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Structural insights into the intermolecular interaction of the adhesin SdrC in the pathogenicity of Staphylococcus aureus.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
3K3O
 
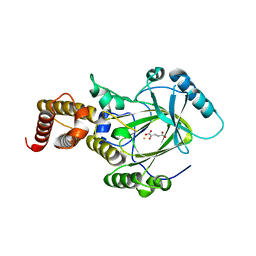 | | Crystal structure of the catalytic core domain of human PHF8 complexed with alpha-ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, FE (II) ION, PHD finger protein 8 | | 著者 | Yu, L, Wang, Y, Huang, S, Wang, J, Deng, Z, Wu, W, Gong, W, Chen, Z. | | 登録日 | 2009-10-03 | | 公開日 | 2010-01-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural insights into a novel histone demethylase PHF8
Cell Res., 20, 2010
|
|
3D4L
 
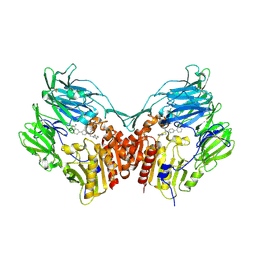 | | Human dipeptidyl peptidase IV/CD26 in complex with a novel inhibitor | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Scapin, G. | | 登録日 | 2008-05-14 | | 公開日 | 2008-07-01 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of new binding elements in DPP-4 inhibition and their applications in novel DPP-4 inhibitor design.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
