4UUT
 
 | |
4USD
 
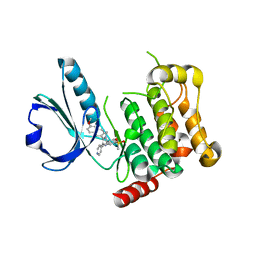 | | Human STK10 (LOK) with SB-633825 | | 分子名称: | 4-{5-(6-methoxynaphthalen-2-yl)-1-methyl-2-[2-methyl-4-(methylsulfonyl)phenyl]-1H-imidazol-4-yl}pyridine, SERINE/THREONINE-PROTEIN KINASE 10 | | 著者 | Elkins, J.M, Salah, E, Szklarz, M, von Delft, F, Canning, P, Raynor, J, Bountra, C, Edwards, A.M, Knapp, S. | | 登録日 | 2014-07-07 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Comprehensive Characterization of the Published Kinase Inhibitor Set.
Nat.Biotechnol., 34, 2016
|
|
4USF
 
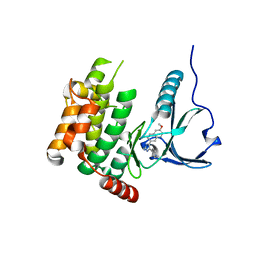 | | Human SLK with SB-440719 | | 分子名称: | 4-[4-(6-methoxynaphthalen-2-yl)-1H-imidazol-5-yl]pyridine, STE20-LIKE SERINE/THREONINE-PROTEIN KINASE | | 著者 | Elkins, J.M, Salah, E, Szklarz, M, von Delft, F, Bountra, C, Edwards, A.M, Knapp, S. | | 登録日 | 2014-07-07 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Comprehensive Characterization of the Published Kinase Inhibitor Set.
Nat.Biotechnol., 34, 2016
|
|
4UT1
 
 | |
2SGQ
 
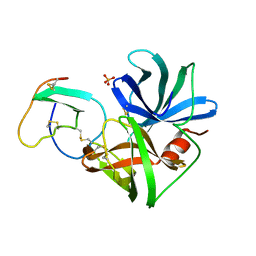 | | GLN 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | 分子名称: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | 著者 | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | 登録日 | 1999-03-25 | | 公開日 | 2003-08-26 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pH's
To be Published
|
|
2RMX
 
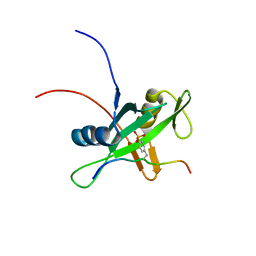 | | Solution structure of the SHP-1 C-terminal SH2 domain complexed with a tyrosine-phosphorylated peptide from NKG2A | | 分子名称: | NKG2-A/NKG2-B type II integral membrane protein, Tyrosine-protein phosphatase non-receptor type 6 | | 著者 | Kasai, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-11-30 | | 公開日 | 2008-12-02 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the recognition of the two NKG2A immunoreceptor tyrosine-based inhibitory motifs (ITIMs) by the C-terminal SH2 domain of protein tyrosine phosphatase SHP-1
To be Published
|
|
4UZR
 
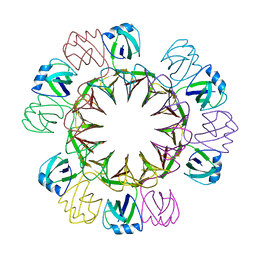 | |
4UXE
 
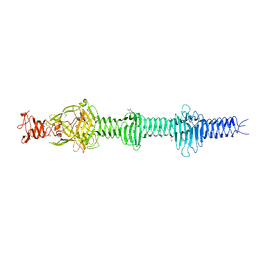 | | Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fibre protein gp34, P21 selenomethionine crystal | | 分子名称: | GLYCEROL, LARGE TAIL FIBER PROTEIN P34 | | 著者 | Granell, M, Alvira, S, Garcia-Doval, C, Singh, A.K, van Raaij, M.J. | | 登録日 | 2014-08-22 | | 公開日 | 2015-08-19 | | 最終更新日 | 2017-07-19 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34.
Viruses, 9, 2017
|
|
2RVB
 
 | |
2RKG
 
 | | HIV-1 PR resistant mutant + LPV | | 分子名称: | GLYCEROL, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, PROTEASE RETROPEPSIN | | 著者 | Rezacova, P, Brynda, J, Kozisek, M, Saskova, K, Konvalinka, J. | | 登録日 | 2007-10-16 | | 公開日 | 2008-08-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Ninety-nine is not enough: molecular characterization of inhibitor-resistant human immunodeficiency virus type 1 protease mutants with insertions in the flap region
J.Virol., 82, 2008
|
|
2RJP
 
 | | Crystal structure of ADAMTS4 with inhibitor bound | | 分子名称: | ADAMTS-4, CALCIUM ION, N-({4'-[(4-isobutyrylphenoxy)methyl]biphenyl-4-yl}sulfonyl)-D-valine, ... | | 著者 | Mosyak, L, Stahl, M, Somers, W. | | 登録日 | 2007-10-15 | | 公開日 | 2007-12-11 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5.
Protein Sci., 17, 2008
|
|
2SGE
 
 | | GLU 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 10.7 | | 分子名称: | Ovomucoid, PHOSPHATE ION, POTASSIUM ION, ... | | 著者 | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | 登録日 | 1999-03-25 | | 公開日 | 2003-08-26 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Water molecules participate in proteinase-inhibitor interactions: crystal structures of Leu18, Ala18, and Gly18 variants of turkey ovomucoid inhibitor third domain complexed with Streptomyces griseus proteinase B.
Protein Sci., 4, 1995
|
|
2RSK
 
 | | RNA aptamer against prion protein in complex with the partial binding peptide | | 分子名称: | RNA (5'-R(*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*A)-3'), partial binding peptide of Major prion protein | | 著者 | Mashima, T, Nishikawa, F, Kamatari, Y.O, Fujiwara, H, Nishikawa, S, Kuwata, K, Katahira, M. | | 登録日 | 2012-03-08 | | 公開日 | 2013-02-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Anti-prion activity of an RNA aptamer and its structural basis
Nucleic Acids Res., 41, 2013
|
|
2RHE
 
 | |
2RQL
 
 | |
4V8S
 
 | | Archaeal RNAP-DNA binary complex at 4.32Ang | | 分子名称: | 5'-D(*AP*TP*AP*GP*AP*GP*TP*AP*TP*AP*AP*GP*AP*TP *AP*G)-3', 5'-D(*TP*CP*TP*TP*AP*TP*AP*CP*TP*CP*TP*AP*TP*CP)-3', DNA-DIRECTED RNA POLYMERASE, ... | | 著者 | Wojtas, M.N, Mogni, M, Millet, O, Bell, S.D, Abrescia, N.G.A. | | 登録日 | 2012-07-12 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (4.323 Å) | | 主引用文献 | Structural and Functional Analyses of the Interaction of Archaeal RNA Polymerase with DNA.
Nucleic Acids Res., 40, 2012
|
|
6L8N
 
 | | Crystal structure of the K. lactis Rad5 | | 分子名称: | DNA repair protein RAD5, ZINC ION | | 著者 | Shen, M, Xiang, S. | | 登録日 | 2019-11-06 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
7S1W
 
 | | The AAVrh.10-glycan complex | | 分子名称: | Capsid protein VP1, beta-D-galactopyranose | | 著者 | Mietzsch, M, McKenna, R. | | 登録日 | 2021-09-02 | | 公開日 | 2021-09-22 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.09 Å) | | 主引用文献 | Structural Study of Aavrh.10 Receptor and Antibody Interactions.
J.Virol., 95, 2021
|
|
6LEB
 
 | | Staphylococcus aureus surface protein SdrC mutant-P366H | | 分子名称: | GLYCEROL, MAGNESIUM ION, Ser-Asp rich fibrinogen-binding, ... | | 著者 | Hang, T, Zhang, M, Wang, J. | | 登録日 | 2019-11-25 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Structural insights into the intermolecular interaction of the adhesin SdrC in the pathogenicity of Staphylococcus aureus.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
6R64
 
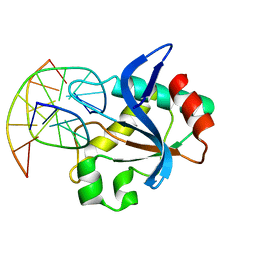 | | N-terminal domain of modification dependent EcoKMcrA restriction endonuclease (NEco) in complex with C5mCGG target sequence | | 分子名称: | 5-methylcytosine-specific restriction enzyme A, DNA (5'-D(*GP*AP*AP*CP*(5CM)P*GP*GP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*CP*(5CM)P*GP*GP*TP*TP*C)-3') | | 著者 | Slyvka, A, Zagorskaite, E, Czapinska, H, Sasnauskas, G, Bochtler, M. | | 登録日 | 2019-03-26 | | 公開日 | 2019-10-23 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Crystal structure of the EcoKMcrA N-terminal domain (NEco): recognition of modified cytosine bases without flipping.
Nucleic Acids Res., 47, 2019
|
|
7YXU
 
 | | Crystal structure of agonistic antibody 1618 fab domain bound to human 4-1BB. | | 分子名称: | MANGANESE (II) ION, Tumor necrosis factor receptor superfamily member 9, heavy chain of Fab, ... | | 著者 | Hakansson, M, Rose, N, Petersson, J, Enell Smith, K, Thorolfsson, M, von Schantz, L. | | 登録日 | 2022-02-16 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | The Bispecific Tumor Antigen-Conditional 4-1BB x 5T4 Agonist, ALG.APV-527, Mediates Strong T-Cell Activation and Potent Antitumor Activity in Preclinical Studies.
Mol.Cancer Ther., 22, 2023
|
|
6L88
 
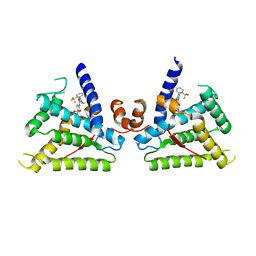 | |
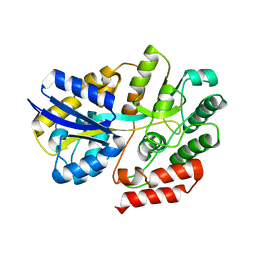
6LCF
 
 | |
7RUU
 
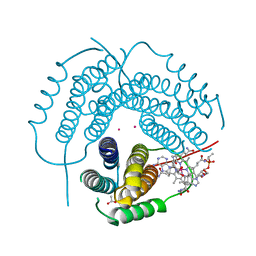 | | Structure of Human ATP:Cobalamin Adenosyltransferase R190C bound to adenosylcobalamin | | 分子名称: | 5'-DEOXYADENOSINE, ACETATE ION, COBALAMIN, ... | | 著者 | Mascarenhas, R, Gouda, H, Koutmos, M, Banerjee, R. | | 登録日 | 2021-08-18 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Patient mutations in human ATP:cob(I)alamin adenosyltransferase differentially affect its catalytic versus chaperone functions.
J.Biol.Chem., 297, 2021
|
|
7RUT
 
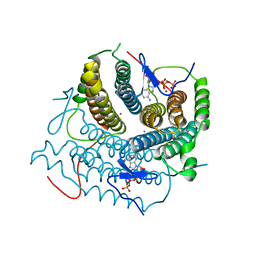 | | Structure of Human ATP:Cobalamin Adenosyltransferase R190C bound to ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Corrinoid adenosyltransferase, GLYCEROL, ... | | 著者 | Mascarenhas, R, Gouda, H, Koutmos, M, Banerjee, R. | | 登録日 | 2021-08-18 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Patient mutations in human ATP:cob(I)alamin adenosyltransferase differentially affect its catalytic versus chaperone functions.
J.Biol.Chem., 297, 2021
|
|
