7Y3B
 
 | | Crystal structure of TRIM7 bound to GN1 | | 分子名称: | E3 ubiquitin-protein ligase TRIM7,TRIM7-GN1 | | 著者 | Dong, C, Yan, X. | | 登録日 | 2022-06-10 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Y3C
 
 | | Crystal structure of TRIM7 bound to RACO-1 | | 分子名称: | E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,TRIM7-RACO-1 | | 著者 | Dong, C, Yan, X. | | 登録日 | 2022-06-10 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8E3N
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with rifamycin S | | 分子名称: | Nuclear receptor subfamily 1 group I member 2, Rifamycin S | | 著者 | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Chen, T. | | 登録日 | 2022-08-17 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5KTU
 
 | | Crystal structure of the bromodomain of human CREBBP bound to pyrazolopiperidine scaffold | | 分子名称: | 1-(3-phenylazanyl-1,4,6,7-tetrahydropyrazolo[4,3-c]pyridin-5-yl)ethanone, CREB-binding protein, DIMETHYL SULFOXIDE | | 著者 | Jayaram, H, Poy, F, Setser, J.W, Bellon, S.F. | | 登録日 | 2016-07-12 | | 公開日 | 2016-11-02 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Discovery of a Potent and Selective in Vivo Probe (GNE-272) for the Bromodomains of CBP/EP300.
J. Med. Chem., 59, 2016
|
|
6S6B
 
 | | Type III-B Cmr-beta Cryo-EM structure of the Apo state | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-07-02 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (2.75 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6S8B
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 1 | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-07-09 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (2.41 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6S91
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 2 | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-07-11 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6SIC
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA | | 分子名称: | CRISPR-associated RAMP protein, Cmr1 family, Cmr4 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-08-09 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6S8E
 
 | | Cryo-EM structure of the type III-B Cmr-beta complex bound to non-cognate target RNA | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-07-09 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6SH8
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 2, in the presence of ssDNA | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-08-06 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6SHB
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 1, in the presence of ssDNA | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-08-06 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
5MES
 
 | | MCL1 FAB COMPLEX IN COMPLEX WITH COMPOUND 29 | | 分子名称: | (5~{R},13~{S},17~{S})-5-[[4-chloranyl-3-(2-phenylethyl)phenyl]methyl]-13-[(4-chlorophenyl)methyl]-8-methyl-1,4,8,12,16-pentazatricyclo[15.8.1.0^{20,25}]hexacosa-20(25),21,23-triene-3,7,15,26-tetrone, Heavy Chain, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog,Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | 著者 | Hargreaves, D. | | 登録日 | 2016-11-16 | | 公開日 | 2017-01-18 | | 最終更新日 | 2017-03-08 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
4IH1
 
 | | Crystal structure of Karrikin Insensitive 2 (KAI2) from Arabidopsis thaliana | | 分子名称: | Hydrolase, alpha/beta fold family protein | | 著者 | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | 登録日 | 2012-12-18 | | 公開日 | 2013-01-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
4IH9
 
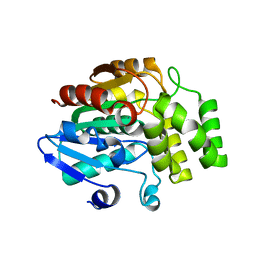 | | Crystal structure of rice DWARF14 (D14) | | 分子名称: | Dwarf 88 esterase | | 著者 | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | 登録日 | 2012-12-18 | | 公開日 | 2013-01-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
4IH4
 
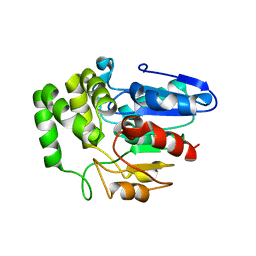 | | Crystal structure of Arabidopsis DWARF14 orthologue, AtD14 | | 分子名称: | AT3g03990/T11I18_10 | | 著者 | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | 登録日 | 2012-12-18 | | 公開日 | 2013-01-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
5MEV
 
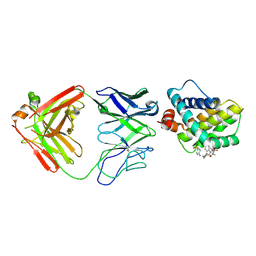 | | MCL1 FAB COMPLEX IN COMPLEX WITH COMPOUND 21 | | 分子名称: | (5~{R},13~{S},17~{S})-5-[(3,4-dichlorophenyl)methyl]-8-methyl-13-[(4-methylsulfonylphenyl)methyl]-1,4,8,12,16-pentazatricyclo[15.8.1.0^{20,25}]hexacosa-20,22,24-triene-3,7,15,26-tetrone, Fab Heavy Chain, Fab Light Chain, ... | | 著者 | Hargreaves, D. | | 登録日 | 2016-11-16 | | 公開日 | 2017-01-18 | | 最終更新日 | 2017-03-08 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors
Acs Med.Chem.Lett., 8, 2017
|
|
6S3D
 
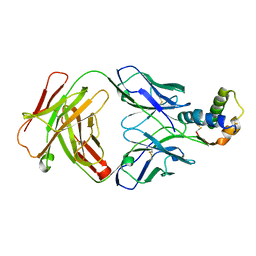 | |
4IHA
 
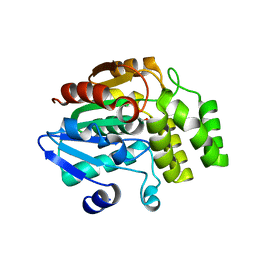 | | Crystal structure of rice DWARF14 (D14) in complex with a GR24 hydrolysis intermediate | | 分子名称: | (2R,3R)-2,4,4-trihydroxy-3-methylbutanal, Dwarf 88 esterase | | 著者 | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | 登録日 | 2012-12-18 | | 公開日 | 2013-01-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
6V2S
 
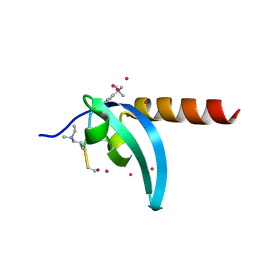 | | Crystal Structure of chromodomain of MPP8 in complex with inhibitor UNC3866 | | 分子名称: | M-phase phosphoprotein 8, UNC3866, UNKNOWN ATOM OR ION | | 著者 | Liu, Y, Tempel, W, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2019-11-25 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
5JB1
 
 | | Pseudo-atomic structure of Human Papillomavirus Type 59 L1 Virus-like Particle | | 分子名称: | Major capsid protein L1 | | 著者 | Li, Z.H, Yan, X.D, Yu, H, Zheng, Q.B, Gu, Y, Li, S.W. | | 登録日 | 2016-04-13 | | 公開日 | 2016-05-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | The C-Terminal Arm of the Human Papillomavirus Major Capsid Protein Is Immunogenic and Involved in Virus-Host Interaction.
Structure, 24, 2016
|
|
5J6R
 
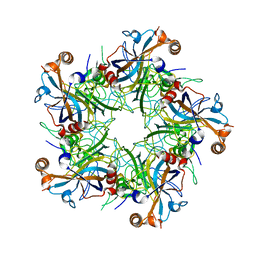 | | Crystal structure of Human Papillomavirus Type 59 L1 pentamer | | 分子名称: | Major capsid protein L1 | | 著者 | Li, Z.H, Yan, X.D, Yu, H, Gu, Y, Li, S.W. | | 登録日 | 2016-04-05 | | 公開日 | 2016-05-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (4.011 Å) | | 主引用文献 | The C-Terminal Arm of the Human Papillomavirus Major Capsid Protein Is Immunogenic and Involved in Virus-Host Interaction.
Structure, 24, 2016
|
|
6V2R
 
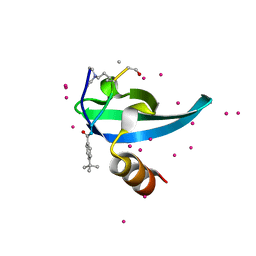 | | Crystal Structure of chromodomain of CBX7 mutant V13A in complex with inhibitor UNC3866 | | 分子名称: | Chromobox protein homolog 7, UNC3866, UNKNOWN ATOM OR ION | | 著者 | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2019-11-25 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V2D
 
 | | Crystal Structure of chromodomain of CDYL2 in complex with inhibitor UNC3866 | | 分子名称: | Chromodomain Y-like protein 2, UNC3866, UNKNOWN ATOM OR ION | | 著者 | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2019-11-22 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
2MXB
 
 | |
2MJ8
 
 | | Solution structure of CDYL2 chromodomain | | 分子名称: | Chromodomain Y-like protein 2 | | 著者 | Qin, S, Houliston, S, Arrowsmith, C.H, Edwards, A.M, Wu, H, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2013-12-28 | | 公開日 | 2014-04-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
