6AGJ
 
 | |
6IFU
 
 | | Cryo-EM structure of type III-A Csm-CTR2-dsDNA complex | | 分子名称: | CTR2, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-21 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IFR
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-NTR, ATP bound | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Type III-A CRISPR-associated RAMP protein Csm3, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-21 | | 公開日 | 2018-12-12 | | 最終更新日 | 2019-01-23 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6SEB
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB in complex with IPTG | | 分子名称: | 1-methylethyl 1-thio-beta-D-galactopyranoside, ACETATE ION, Beta-galactosidase, ... | | 著者 | Rutkiewicz, M, Bujacz, A, Kaminska, P, Bujacz, G. | | 登録日 | 2019-07-29 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.272 Å) | | 主引用文献 | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
7C8P
 
 | |
7CKM
 
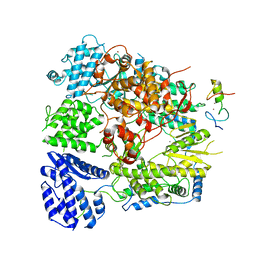 | | Structure of Machupo virus polymerase bound to Z matrix protein (monomeric complex) | | 分子名称: | MANGANESE (II) ION, RING finger protein Z, RNA-directed RNA polymerase L, ... | | 著者 | Xu, X, Peng, R, Peng, Q, Shi, Y. | | 登録日 | 2020-07-17 | | 公開日 | 2021-05-05 | | 最終更新日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals
Nat Microbiol, 6, 2021
|
|
7CKL
 
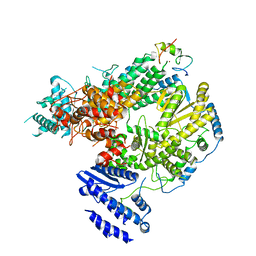 | | Structure of Lassa virus polymerase bound to Z matrix protein | | 分子名称: | MANGANESE (II) ION, RING finger protein Z, RNA-directed RNA polymerase L, ... | | 著者 | Xu, X, Peng, R, Peng, Q, Shi, Y. | | 登録日 | 2020-07-17 | | 公開日 | 2021-05-05 | | 最終更新日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals
Nat Microbiol, 6, 2021
|
|
6SED
 
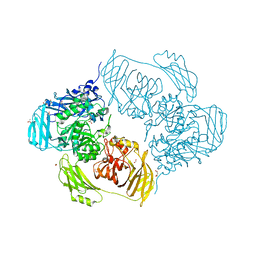 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB in complex with galactose | | 分子名称: | ACETATE ION, Beta-galactosidase, FORMIC ACID, ... | | 著者 | Rutkiewicz, M, Bujacz, A, Kaminska, P, Bujacz, G. | | 登録日 | 2019-07-29 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.233 Å) | | 主引用文献 | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
6AGI
 
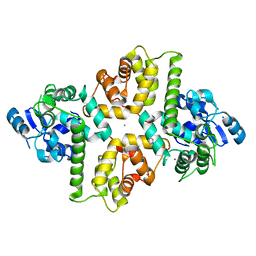 | | Crystal Structure of EFHA2 in Ca-binding State | | 分子名称: | CALCIUM ION, Calcium uptake protein 3, mitochondrial, ... | | 著者 | Yangfei, X, Xue, Y, Yuequan, S. | | 登録日 | 2018-08-11 | | 公開日 | 2019-01-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.799 Å) | | 主引用文献 | Dimerization of MICU Proteins Controls Ca2+Influx through the Mitochondrial Ca2+Uniporter.
Cell Rep, 26, 2019
|
|
6KDV
 
 | | Crystal structure of TtCas1-DNA complex | | 分子名称: | CRISPR-associated endonuclease Cas1 2, DNA (5'-D(*GP*AP*GP*TP*CP*GP*AP*TP*GP*CP*TP*GP*GP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*CP*CP*AP*GP*CP*AP*TP*CP*GP*AP*CP*TP*C)-3') | | 著者 | Wang, Y.L, Li, J.Z, Yang, J, Wang, J.Y. | | 登録日 | 2019-07-02 | | 公開日 | 2019-12-18 | | 最終更新日 | 2020-05-06 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Crystal structure of Cas1 in complex with branched DNA.
Sci China Life Sci, 63, 2020
|
|
6JE3
 
 | | Crystal structure of Nme2Cas9 in complex with sgRNA and target DNA (AGGCCC PAM) with 5 nt overhang | | 分子名称: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, non-target DNA strand, ... | | 著者 | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | 登録日 | 2019-02-03 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.931 Å) | | 主引用文献 | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JDV
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in catalytic state | | 分子名称: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, MAGNESIUM ION, ... | | 著者 | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | 登録日 | 2019-02-02 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
7F0M
 
 | | Crystal Structure of human Pin1 complexed with a potent covalent inhibitor | | 分子名称: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 8-(2-chloranylethanoyl)-4-[(5-naphthalen-1-ylfuran-2-yl)methyl]-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Liu, L, Li, J. | | 登録日 | 2021-06-05 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
3NJY
 
 | | Crystal structure of JMJD2A complexed with 5-carboxy-8-hydroxyquinoline | | 分子名称: | 8-hydroxyquinoline-5-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | 著者 | King, O.N.F, Clifton, I.J, Wang, M, Maloney, D.J, Jadhav, A, Oppermann, U, Heightman, T.D, Simeonov, A, McDonough, M.A, Schofield, C.J. | | 登録日 | 2010-06-18 | | 公開日 | 2010-12-08 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors
Plos One, 5, 2010
|
|
7K6Q
 
 | | Active state Dot1 bound to the H4K16ac nucleosome | | 分子名称: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Valencia-Sanchez, M.I, De Ioannes, P.E, Miao, W, Truong, D.M, Lee, R, Armache, J.-P, Boeke, J.D, Armache, K.-J. | | 登録日 | 2020-09-21 | | 公開日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Regulation of the Dot1 histone H3K79 methyltransferase by histone H4K16 acetylation.
Science, 371, 2021
|
|
7K6P
 
 | | Active state Dot1 bound to the unacetylated H4 nucleosome | | 分子名称: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Valencia-Sanchez, M.I, De Ioannes, P.E, Miao, W, Truong, D.M, Lee, R, Armache, J.-P, Boeke, J.D, Armache, K.-J. | | 登録日 | 2020-09-21 | | 公開日 | 2021-02-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Regulation of the Dot1 histone H3K79 methyltransferase by histone H4K16 acetylation.
Science, 371, 2021
|
|
7JZV
 
 | | Cryo-EM structure of the BRCA1-UbcH5c/BARD1 E3-E2 module bound to a nucleosome | | 分子名称: | BRCA1,Ubiquitin-conjugating enzyme E2 D3, BRCA1-associated RING domain protein 1, Histone H2A type 2-A, ... | | 著者 | Witus, S.R, Burrell, A.L, Hansen, J.M, Farrell, D.P, Dimaio, F, Kollman, J.M, Klevit, R.E. | | 登録日 | 2020-09-02 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | BRCA1/BARD1 site-specific ubiquitylation of nucleosomal H2A is directed by BARD1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6IK4
 
 | | A Novel M23 Metalloprotease Pseudoalterin from Deep-sea | | 分子名称: | Elastinolytic metalloprotease, GLYCEROL, ZINC ION | | 著者 | Zhao, H.L, Tang, B.L, Yang, J, Chen, X.L, Zhang, Y.Z. | | 登録日 | 2018-10-14 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A predator-prey interaction between a marine Pseudoalteromonas sp. and Gram-positive bacteria.
Nat Commun, 11, 2020
|
|
7DTE
 
 | | SARS-CoV-2 RdRP catalytic complex with T33-1 RNA | | 分子名称: | Non-structural protein 7, Non-structural protein 8, RNA (33-MER), ... | | 著者 | Wang, Q, Gong, P. | | 登録日 | 2021-01-04 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Remdesivir overcomes the S861 roadblock in SARS-CoV-2 polymerase elongation complex.
Cell Rep, 37, 2021
|
|
5EUV
 
 | | Crystal structure of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d strain | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Beta-D-galactosidase, ... | | 著者 | Rutkiewicz-Krotewicz, M, Bujacz, A, Pietrzyk, A.J, Sekula, B, Bujacz, G. | | 登録日 | 2015-11-19 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural studies of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7E1L
 
 | | Crystal structure of apo form PhlH | | 分子名称: | DUF1956 domain-containing protein | | 著者 | Zhang, N, Wu, J, He, Y.X, Ge, H. | | 登録日 | 2021-02-01 | | 公開日 | 2022-02-02 | | 最終更新日 | 2022-08-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular basis for coordinating secondary metabolite production by bacterial and plant signaling molecules.
J.Biol.Chem., 298, 2022
|
|
7E1N
 
 | | Crystal structure of PhlH in complex with 2,4-diacetylphloroglucinol | | 分子名称: | 2,4-bis[(1R)-1-oxidanylethyl]benzene-1,3,5-triol, DUF1956 domain-containing protein | | 著者 | Zhang, N, Wu, J, He, Y.X, Ge, H. | | 登録日 | 2021-02-02 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Molecular basis for coordinating secondary metabolite production by bacterial and plant signaling molecules.
J.Biol.Chem., 298, 2022
|
|
8GS9
 
 | | SARS-CoV-2 BA.2 spike RBD in complex bound with VacBB-551 | | 分子名称: | Heavy chain of VacBB-551, Light chain of VacBB-551, Spike glycoprotein | | 著者 | Liu, C.C, Ju, B, Shen, S.L, Zhang, Z. | | 登録日 | 2022-09-05 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Omicron BQ.1.1 and XBB.1 unprecedentedly escape broadly neutralizing antibodies elicited by prototype vaccination.
Cell Rep, 42, 2023
|
|
3PP6
 
 | | REP1-NXSQ fatty acid transporter Y128F mutant | | 分子名称: | PALMITOLEIC ACID, ReP1-NCXSQ | | 著者 | Berberian, G, Bollo, M, Howard, E, Cousido-Siah, A, Mitschler, A, Ayoub, D, Sanglier-Cianferani, S, Van Dorsselaer, A, DiPolo, R, Beauge, L, Petrova, T, Schulze-Briese, C, Wang, M, Podjarny, A. | | 登録日 | 2010-11-24 | | 公開日 | 2011-12-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and functional studies of ReP1-NCXSQ, a protein regulating the squid nerve Na+/Ca2+ exchanger.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3PPT
 
 | | REP1-NXSQ fatty acid transporter | | 分子名称: | PALMITOLEIC ACID, ReP1-NCXSQ | | 著者 | Berberian, G, Bollo, M, Howard, E, Cousido-Siah, A, Mitschler, A, Ayoub, D, Sanglier-Cianferani, S, Van Dorsselaer, A, DiPolo, R, Beauge, L, Petrova, T, Schulze-Briese, C, Wang, M, Podjarny, A. | | 登録日 | 2010-11-25 | | 公開日 | 2011-12-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Structural and functional studies of ReP1-NCXSQ, a protein regulating the squid nerve Na+/Ca2+ exchanger.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
