4RS9
 
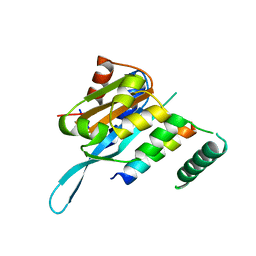 | | Structure of Myc3 N-terminal JAZ-binding domain [44-238] in complex with Jas motif of JAZ9 | | 分子名称: | Protein TIFY 7, Transcription factor MYC3 | | 著者 | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J.S, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | 登録日 | 2014-11-07 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
4RQW
 
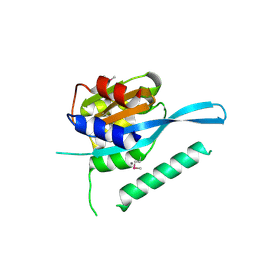 | | Crystal structure of Myc3 N-terminal JAZ-binding domain [44-238] from Arabidopsis | | 分子名称: | CALCIUM ION, Transcription factor MYC3 | | 著者 | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | 登録日 | 2014-11-05 | | 公開日 | 2015-08-12 | | 最終更新日 | 2015-09-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
8TB0
 
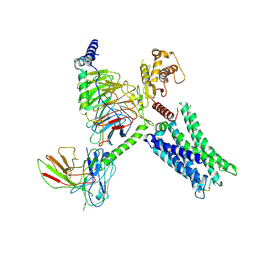 | | Cryo-EM Structure of GPR61-G protein complex stabilized by scFv16 | | 分子名称: | GPR61 fused to dominant negative G alpha S/I N18 chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Lees, J.A, Dias, J.M, Han, S. | | 登録日 | 2023-06-28 | | 公開日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | An inverse agonist of orphan receptor GPR61 acts by a G protein-competitive allosteric mechanism.
Nat Commun, 14, 2023
|
|
8TB7
 
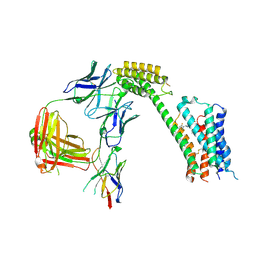 | | Cryo-EM Structure of GPR61- | | 分子名称: | 6-{[(3,5-difluoropyridin-4-yl)methyl]amino}-N-(4-ethoxy-6-methylpyrimidin-2-yl)-2-methoxy-N-(2-methoxyethyl)pyridine-3-sulfonamide, Fab hinge-binding nanobody, Fab24 BAK5 heavy chain, ... | | 著者 | Lees, J.A, Dias, J.M, Han, S. | | 登録日 | 2023-06-28 | | 公開日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | An inverse agonist of orphan receptor GPR61 acts by a G protein-competitive allosteric mechanism.
Nat Commun, 14, 2023
|
|
6KZC
 
 | | crystal structure of TRKc in complex with 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methyl-N-(3-((4- methylpiperazin-1-yl)methyl)-5- (trifluoromethyl)phenyl)benzamide | | 分子名称: | 3-(2-imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methyl-~{N}-[3-(4-methylpiperazin-1-yl)-5-(trifluoromethyl)phenyl]benzamide, NT-3 growth factor receptor | | 著者 | Zhang, Z.M, Wang, Y. | | 登録日 | 2019-09-23 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Design, synthesis and biological evaluation of 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methylbenzamides as potent and selective pan-tropomyosin receptor kinase (TRK) inhibitors.
Eur.J.Med.Chem., 179, 2019
|
|
8P0C
 
 | |
8P0E
 
 | |
3ROQ
 
 | | Crystal structure of human CD38 in complex with compound CZ-46 | | 分子名称: | (2R,3R,4S,5S)-4-fluoro-3,5-dihydroxytetrahydrofuran-2-yl 2-phenylethyl hydrogen (S)-phosphate, ADP-ribosyl cyclase 1 | | 著者 | Zhang, H, Lee, H.C, Hao, Q. | | 登録日 | 2011-04-26 | | 公開日 | 2011-12-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Catalysis-based inhibitors of the calcium signaling function of CD38.
Biochemistry, 51, 2012
|
|
4XXI
 
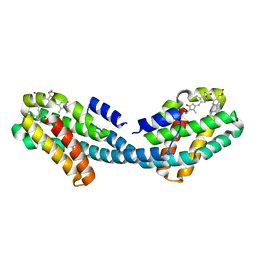 | | Crystal structure of the Bilin-binding domain of phycobilisome core-membrane linker ApcE | | 分子名称: | PHYCOCYANOBILIN, Phycobiliprotein ApcE | | 著者 | Tang, K, Ding, W.-L, Hoppner, A, Gartner, W, Zhao, K.-H. | | 登録日 | 2015-01-30 | | 公開日 | 2015-12-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The terminal phycobilisome emitter, LCM: A light-harvesting pigment with a phytochrome chromophore
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3ROM
 
 | |
4XXU
 
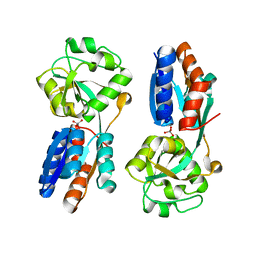 | | ModA - chromate bound | | 分子名称: | Chromate, Molybdate-binding periplasmic protein | | 著者 | He, C, Karpus, J, Ajiboye, I. | | 登録日 | 2015-01-30 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Structure of chromate bound ModA from E.coli at 1.43 Angstroms resolution.
To Be Published
|
|
6KZD
 
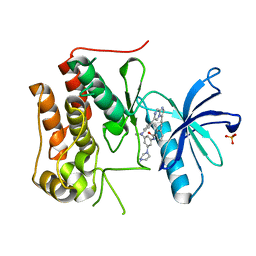 | | Crystal structure of TRKc in complex with 3-((6-(4-aminophenyl)imidazo[1,2-a]pyrazin-3-yl)ethynyl)- N-(3-isopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-2- methylbenzamide | | 分子名称: | 3-[2-[6-(4-aminophenyl)imidazo[1,2-a]pyrazin-3-yl]ethynyl]-2-methyl-~{N}-[3-(4-methylpiperazin-1-yl)-5-propan-2-yl-phenyl]benzamide, NT-3 growth factor receptor, PHOSPHATE ION | | 著者 | Wang, Y, Zhang, Z.M. | | 登録日 | 2019-09-23 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.708 Å) | | 主引用文献 | Design, synthesis and biological evaluation of 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methylbenzamides as potent and selective pan-tropomyosin receptor kinase (TRK) inhibitors.
Eur.J.Med.Chem., 179, 2019
|
|
4XXK
 
 | | Crystal structure of the Semet-derivative of the Bilin-binding domain of phycobilisome core-membrane linker ApcE | | 分子名称: | PHYCOCYANOBILIN, Phycobiliprotein ApcE | | 著者 | Tang, K, Ding, W.-L, Hoppner, A, Gartner, W, Zhao, K.-H. | | 登録日 | 2015-01-30 | | 公開日 | 2015-12-16 | | 最終更新日 | 2016-01-13 | | 実験手法 | X-RAY DIFFRACTION (2.97 Å) | | 主引用文献 | The terminal phycobilisome emitter, LCM: A light-harvesting pigment with a phytochrome chromophore.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1ORD
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF A PLP-DEPENDENT ORNITHINE DECARBOXYLASE FROM LACTOBACILLUS 30A TO 3.1 ANGSTROMS RESOLUTION | | 分子名称: | ORNITHINE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Hackert, M.L, Momany, C, Ernst, S, Ghosh, R. | | 登録日 | 1995-02-08 | | 公開日 | 1995-09-15 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystallographic structure of a PLP-dependent ornithine decarboxylase from Lactobacillus 30a to 3.0 A resolution.
J.Mol.Biol., 252, 1995
|
|
7E3X
 
 | | Crystal structure of SDR family NAD(P)-dependent oxidoreductase from exiguobacterium | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase | | 著者 | Chen, L, Tang, J, Yuan, S, Zhang, F, Chen, S. | | 登録日 | 2021-02-09 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Structure-guided evolution of a ketoreductase forefficient and stereoselective bioreduction of bulkyalpha-aminobeta-keto esters
Catalysis Science And Technology, 2021
|
|
8PY3
 
 | | Crystal structure of human Sirt2 in complex with a 1,2,4-oxadiazole based inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-chloranyl-~{N}-[4-[5-[[(3~{S})-1-[(3-fluoranyl-2-methyl-phenyl)methyl]piperidin-3-yl]methyl]-1,2,4-oxadiazol-3-yl]phenyl]benzamide, ... | | 著者 | Friedrich, F, Colcerasa, A, Einsle, O, Jung, M. | | 登録日 | 2023-07-24 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structure-Activity Studies of 1,2,4-Oxadiazoles for the Inhibition of the NAD + -Dependent Lysine Deacylase Sirtuin 2.
J.Med.Chem., 67, 2024
|
|
8QJ4
 
 | |
8QJ3
 
 | |
4WN9
 
 | | Structure of the Nitrogenase MoFe Protein from Clostridium pasteurianum Pressurized with Xenon | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | 著者 | Morrison, C.N, Hoy, J.A, Rees, D.C. | | 登録日 | 2014-10-11 | | 公開日 | 2015-03-18 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Substrate Pathways in the Nitrogenase MoFe Protein by Experimental Identification of Small Molecule Binding Sites.
Biochemistry, 54, 2015
|
|
4YLA
 
 | | Crystal structure of the indole prenyltransferase MpnD complexed with indolactam V and DMSPP | | 分子名称: | (2S,5S)-5-(hydroxymethyl)-1-methyl-2-(propan-2-yl)-1,2,4,5,6,8-hexahydro-3H-[1,4]diazonino[7,6,5-cd]indol-3-one, Aromatic prenyltransferase, DIMETHYLALLYL S-THIOLODIPHOSPHATE, ... | | 著者 | Mori, T, Morita, H, Abe, I. | | 登録日 | 2015-03-05 | | 公開日 | 2016-03-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Manipulation of prenylation reactions by structure-based engineering of bacterial indolactam prenyltransferases.
Nat Commun, 7, 2016
|
|
4PSX
 
 | | Crystal structure of histone acetyltransferase complex | | 分子名称: | COENZYME A, Histone H3, Histone H4, ... | | 著者 | Yang, M, Li, Y. | | 登録日 | 2014-03-08 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.509 Å) | | 主引用文献 | Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex
Genes Dev., 28, 2014
|
|
3WJ7
 
 | | Crystal structure of gox2253 | | 分子名称: | MERCURY (II) ION, Putative oxidoreductase | | 著者 | Yuan, Y.A, Yin, B. | | 登録日 | 2013-10-05 | | 公開日 | 2014-06-04 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural insights into substrate and coenzyme preference by SDR family protein Gox2253 from Gluconobater oxydans.
Proteins, 82, 2014
|
|
4YL7
 
 | |
3UCS
 
 | |
8G9V
 
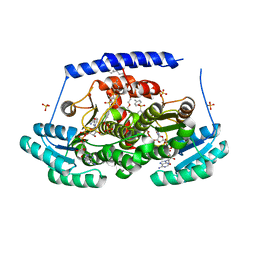 | | Crystal structures of 17-beta-hydroxysteroid dehydrogenase 13 | | 分子名称: | 17-beta-hydroxysteroid dehydrogenase 13, 4-{[2,5-dimethyl-3-(4-methylbenzene-1-sulfonyl)benzene-1-sulfonyl]amino}benzoic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Liu, S. | | 登録日 | 2023-02-22 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.645 Å) | | 主引用文献 | Structural basis of lipid-droplet localization of 17-beta-hydroxysteroid dehydrogenase 13.
Nat Commun, 14, 2023
|
|
