8UP7
 
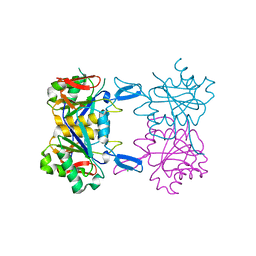 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant K19A) | | 分子名称: | Asparaginase, CHLORIDE ION | | 著者 | Lubkowski, J, Wlodawer, A, Zhang, D. | | 登録日 | 2023-10-21 | | 公開日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UP8
 
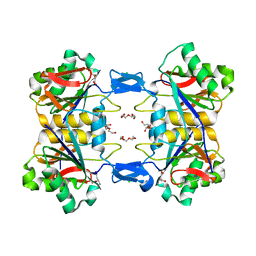 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant Y21F, complex with L-Asp) | | 分子名称: | ASPARTIC ACID, Asparaginase, CHLORIDE ION, ... | | 著者 | Lubkowski, J, Wlodawer, A, Zhang, D. | | 登録日 | 2023-10-21 | | 公開日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UOW
 
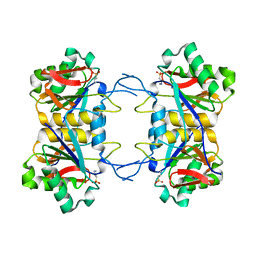 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant Y21A) | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Asparaginase | | 著者 | Lubkowski, J, Wlodawer, A, Zhang, D. | | 登録日 | 2023-10-20 | | 公開日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UP3
 
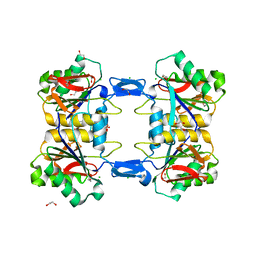 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant Y21F) | | 分子名称: | 1,2-ETHANEDIOL, ASPARTIC ACID, Asparaginase, ... | | 著者 | Lubkowski, J, Wlodawer, A, Zhang, D. | | 登録日 | 2023-10-20 | | 公開日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
2MRO
 
 | |
2MR9
 
 | |
8P3Z
 
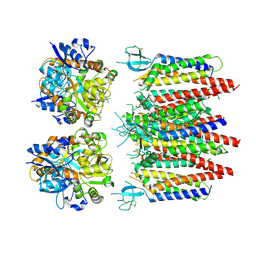 | | Homomeric GluA2 flip R/G-edited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 2 | | 分子名称: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | 著者 | Krieger, J.M, Zhang, D, Yamashita, K, Greger, I.H. | | 登録日 | 2023-05-18 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3Y
 
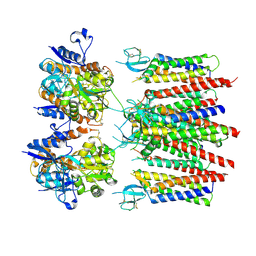 | | Homomeric GluA2 flip R/G-edited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 3 | | 分子名称: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | 著者 | Krieger, J.M, Zhang, D, Yamashita, K, Greger, I.H. | | 登録日 | 2023-05-18 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.55 Å) | | 主引用文献 | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3X
 
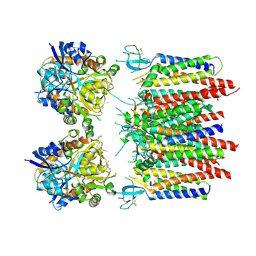 | | Homomeric GluA2 flip R/G-edited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 1 | | 分子名称: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | 著者 | Krieger, J.M, Zhang, D, Yamashita, K, Greger, I.H. | | 登録日 | 2023-05-18 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.36 Å) | | 主引用文献 | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
3NS8
 
 | |
4YH4
 
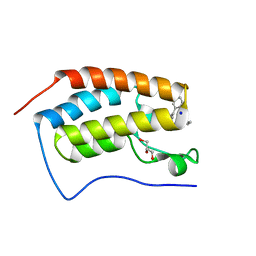 | | Crystal structure of human BRD4(1) in complex with 4-[(5-phenylpyridin-3-yl)carbonyl]-3,4-dihydroquinoxalin-2(1H)-one (compound 19d) | | 分子名称: | 4-[(5-phenylpyridin-3-yl)carbonyl]-3,4-dihydroquinoxalin-2(1H)-one, Bromodomain-containing protein 4, GLYCEROL, ... | | 著者 | Lakshminarasimhan, D, White, A, Suto, R.K. | | 登録日 | 2015-02-26 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Discovery of a new chemical series of BRD4(1) inhibitors using protein-ligand docking and structure-guided design.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
8UK6
 
 | |
1H6H
 
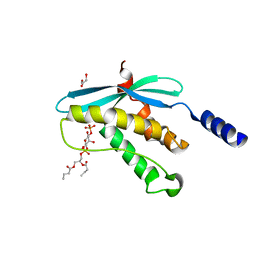 | | Structure of the PX domain from p40phox bound to phosphatidylinositol 3-phosphate | | 分子名称: | 2-(BUTANOYLOXY)-1-{[(HYDROXY{[2,3,4,6-TETRAHYDROXY-5-(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]METHYL}ETHYL BUTANOATE, GLYCEROL, NEUTROPHIL CYTOSOL FACTOR 4 | | 著者 | Karathanassis, D, Bravo, J, Pacold, M, Perisic, O, Williams, R.L. | | 登録日 | 2001-06-15 | | 公開日 | 2001-11-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Crystal Structure of the Px Domain from P40Phox Bound to Phosphatidylinositol 3-Phosphate
Mol.Cell, 8, 2001
|
|
2K57
 
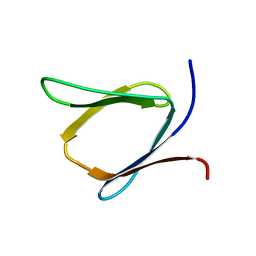 | | Solution NMR Structure of Putative Lipoprotein from Pseudomonas syringae Gene Locus PSPTO2350. Northeast Structural Genomics Target PsR76A. | | 分子名称: | Putative Lipoprotein | | 著者 | Hang, D, Aramini, J.A, Rossi, P, Wang, D, Jiang, M, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-06-25 | | 公開日 | 2008-09-30 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of Putative Lipoprotein from
Pseudomonas syringae Gene Locus PSPTO2350. Northeast Structural Genomics Target PsR76A.
To be Published
|
|
2IWL
 
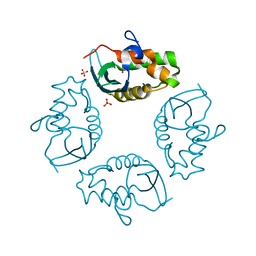 | |
4CQO
 
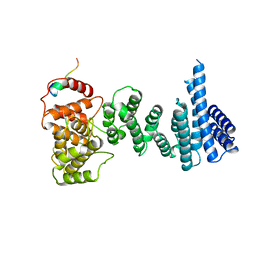 | | Structure of the human CNOT1 superfamily homology domain in complex with a Nanos1 peptide | | 分子名称: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, NANOS HOMOLOG 1 | | 著者 | Raisch, T, Jonas, S, Weichenrieder, O, Bhandari, D, Izaurralde, E. | | 登録日 | 2014-02-21 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Basis for the Nanos-Mediated Recruitment of the Ccr4-not Complex and Translational Repression
Genes Dev., 28, 2014
|
|
8Y0F
 
 | |
8X4F
 
 | |
8X1V
 
 | |
6PLF
 
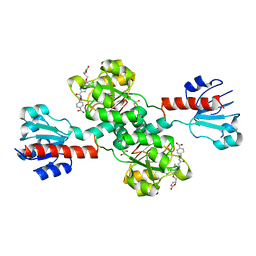 | | Crystal structure of human PHGDH complexed with Compound 1 | | 分子名称: | 1,2-ETHANEDIOL, 4-{(1S)-1-[(5-chloro-6-{[(5S)-2-oxo-1,3-oxazolidin-5-yl]methoxy}-1H-indole-2-carbonyl)amino]-2-hydroxyethyl}benzoic acid, D-3-phosphoglycerate dehydrogenase | | 著者 | Olland, A, Lakshminarasimhan, D, White, A, Suto, R.K. | | 登録日 | 2019-06-30 | | 公開日 | 2019-07-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Inhibition of 3-phosphoglycerate dehydrogenase (PHGDH) by indole amides abrogates de novo serine synthesis in cancer cells.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
4RIM
 
 | |
4RIP
 
 | |
2I9T
 
 | | Structure of NF-kB p65-p50 heterodimer bound to PRDII element of B-interferon promoter | | 分子名称: | 5'-D(*AP*GP*TP*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*CP*TP*G)-3', 5'-D(*CP*AP*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*CP*AP*CP*T)-3', Nuclear factor NF-kappa-B p105 subunit, ... | | 著者 | Escalante, C.R, Shen, L, Thanos, D, Aggarwal, A.K. | | 登録日 | 2006-09-06 | | 公開日 | 2007-02-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of NF-kappaB p50/p65 heterodimer bound to the PRDII DNA element from the interferon-beta promoter
Structure, 10, 2002
|
|
6IS9
 
 | |
6J23
 
 | | Crystal structure of arabidopsis ADAL complexed with GMP | | 分子名称: | Adenosine/AMP deaminase family protein, GUANOSINE-5'-MONOPHOSPHATE, ZINC ION | | 著者 | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | 登録日 | 2018-12-30 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
