6JJI
 
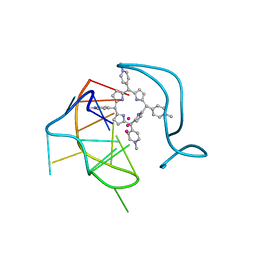 | | Crystal structure of a two-quartet RNA parallel G-quadruplex complexed with the porphyrin TMPyP4 (1:1) | | 分子名称: | (1Z,4Z,9Z,15Z)-5,10,15,20-tetrakis(1-methylpyridin-1-ium-4-yl)-21,23-dihydroporphyrin, POTASSIUM ION, RNA (5'-R(*GP*GP*CP*UP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A)-3') | | 著者 | Zhang, Y.S, Parkinson, G.N, Wei, D.G. | | 登録日 | 2019-02-25 | | 公開日 | 2020-02-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
6JJH
 
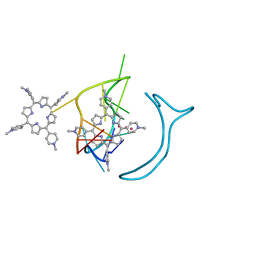 | | Crystal structure of a two-quartet RNA parallel G-quadruplex complexed with the porphyrin TMPyP4 | | 分子名称: | (1Z,4Z,9Z,15Z)-5,10,15,20-tetrakis(1-methylpyridin-1-ium-4-yl)-21,23-dihydroporphyrin, POTASSIUM ION, RNA (5'-R(*GP*GP*CP*UP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A)-3') | | 著者 | Zhang, Y.S, EI Omari, K, Duman, R, Wagner, A, Parkinson, G.N, Wei, D.G. | | 登録日 | 2019-02-25 | | 公開日 | 2020-02-26 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
6JJF
 
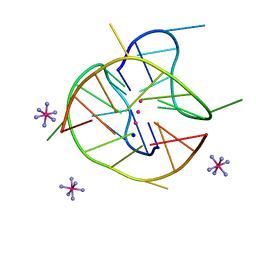 | | Crystal structure of a two-quartet DNA mixed-parallel/antiparallel G-quadruplex | | 分子名称: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*GP*CP*TP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A)-3'), POTASSIUM ION, ... | | 著者 | Zhang, Y.S, EI Omari, K, Duman, R, Wagner, A, Parkinson, G.N, Wei, D.G. | | 登録日 | 2019-02-25 | | 公開日 | 2020-02-26 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
3CE3
 
 | |
3WUY
 
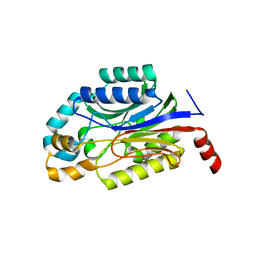 | | Crystal structure of Nit6803 | | 分子名称: | Nitrilase | | 著者 | Yuan, Y.A, Yin, B, Wang, C. | | 登録日 | 2014-05-09 | | 公開日 | 2014-12-31 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural insights into enzymatic activity and substrate specificity determination by a single amino acid in nitrilase from Syechocystis sp. PCC6803
J.Struct.Biol., 188, 2014
|
|
7KXD
 
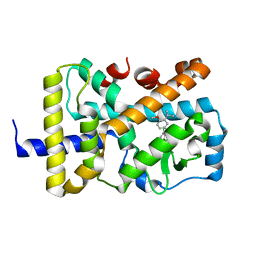 | |
7KXE
 
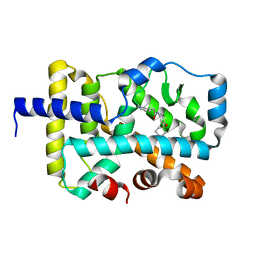 | |
7KXF
 
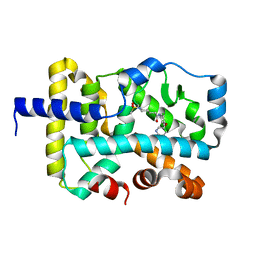 | |
7DFG
 
 | |
7DFH
 
 | |
7DOK
 
 | |
7DOI
 
 | |
3AWD
 
 | | Crystal structure of gox2181 | | 分子名称: | CADMIUM ION, MAGNESIUM ION, Putative polyol dehydrogenase | | 著者 | Adam Yuan, Y, Yuan, Z. | | 登録日 | 2011-03-18 | | 公開日 | 2012-02-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Biochemical and structural analysis of Gox2181, a new member of the SDR superfamily from Gluconobacter oxydans.
Biochem.Biophys.Res.Commun., 415, 2011
|
|
3CTJ
 
 | |
3C1X
 
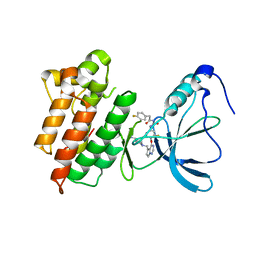 | | Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-MET in complex with a Pyrrolotriazine based inhibitor | | 分子名称: | Hepatocyte growth factor receptor, N-{[4-({5-[(4-aminopiperidin-1-yl)methyl]pyrrolo[2,1-f][1,2,4]triazin-4-yl}oxy)-3-fluorophenyl]carbamoyl}-2-(4-fluorophenyl)acetamide | | 著者 | Sack, J. | | 登録日 | 2008-01-24 | | 公開日 | 2008-03-18 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Identification of pyrrolo[2,1-f][1,2,4]triazine-based inhibitors of Met kinase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CTH
 
 | |
5XXY
 
 | |
7YP1
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 10E4 (localized refinement) | | 分子名称: | 10E4 heavy chain, 10E4 light chain, EBV gH, ... | | 著者 | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | 登録日 | 2022-08-02 | | 公開日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
7YOY
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAbs 3E8 and 5E3 (localized refinement) | | 分子名称: | 3E8 heavy chain, 3E8 light chain, 5E3 heavy chain, ... | | 著者 | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | 登録日 | 2022-08-02 | | 公開日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
7YP2
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 6H2 (localized refinement) | | 分子名称: | 6H2 heavy chain, 6H2 light chain, Envelope glycoprotein H | | 著者 | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | 登録日 | 2022-08-02 | | 公開日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
7TP3
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody K288.2 | | 分子名称: | CACODYLATE ION, K288.2 heavy chain, K288.2 light chain, ... | | 著者 | Yuan, M, Zhu, X, Wilson, I.A. | | 登録日 | 2022-01-24 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques.
Sci Transl Med, 14, 2022
|
|
7TP4
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody K398.22 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, K398.22 heavy chain, ... | | 著者 | Yuan, M, Zhu, X, Wilson, I.A. | | 登録日 | 2022-01-24 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques.
Sci Transl Med, 14, 2022
|
|
6SWY
 
 | | Structure of active GID E3 ubiquitin ligase complex minus Gid2 and delta Gid9 RING domain | | 分子名称: | Glucose-induced degradation protein 8, Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10, Vacuolar import and degradation protein 24, ... | | 著者 | Qiao, S, Prabu, J.R, Schulman, B.A. | | 登録日 | 2019-09-24 | | 公開日 | 2019-11-20 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Interconversion between Anticipatory and Active GID E3 Ubiquitin Ligase Conformations via Metabolically Driven Substrate Receptor Assembly
Mol.Cell, 77, 2020
|
|
8B0K
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli (Apo form) | | 分子名称: | Apolipoprotein N-acyltransferase | | 著者 | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | 登録日 | 2022-09-07 | | 公開日 | 2023-07-12 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0P
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Pam3 | | 分子名称: | Apolipoprotein N-acyltransferase, Pam3-SKKKK, [(2~{S})-3-[(2~{S})-3-azanyl-2-(hexadecanoylamino)-3-oxidanylidene-propyl]sulfanyl-2-hexadecanoyloxy-propyl] hexadecanoate | | 著者 | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | 登録日 | 2022-09-07 | | 公開日 | 2023-07-12 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
