4YVX
 
 | | Crystal structure of AKR1C3 complexed with glimepiride | | 分子名称: | 3-ethyl-4-methyl-N-[2-(4-{[(cis-4-methylcyclohexyl)carbamoyl]sulfamoyl}phenyl)ethyl]-2-oxo-2,5-dihydro-1H-pyrrole-1-car boxamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | 登録日 | 2015-03-20 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
7CFS
 
 | | Cryo-EM strucutre of human acid-sensing ion channel 1a at pH 8.0 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, CHOLESTEROL HEMISUCCINATE, ... | | 著者 | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | 登録日 | 2020-06-28 | | 公開日 | 2020-10-21 | | 実験手法 | ELECTRON MICROSCOPY (3.56 Å) | | 主引用文献 | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
4YVP
 
 | | Crystal Structure of AKR1C1 complexed with glibenclamide | | 分子名称: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | 登録日 | 2015-03-20 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
7CCE
 
 | |
7CFT
 
 | | Cryo-EM strucutre of human acid-sensing ion channel 1a in complex with snake toxin Mambalgin1 at pH 8.0 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, Mambalgin-1 | | 著者 | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | 登録日 | 2020-06-28 | | 公開日 | 2020-10-21 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
4YZ6
 
 | | Crystal Structure of Myc3[44-238] from Arabidopsis in complex with Jaz1 peptide [200-221] | | 分子名称: | Protein TIFY 10A, Transcription factor MYC3 | | 著者 | Ke, J, Zhang, F, Brunzelle, J, Xu, H.E, Melcher, K, HE, S.Y. | | 登録日 | 2015-03-24 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
7X08
 
 | | S protein of SARS-CoV-2 in complex with 2G1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | 著者 | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | 登録日 | 2022-02-21 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Broad ultra-potent neutralization of SARS-CoV-2 variants by monoclonal antibodies specific to the tip of RBD.
Cell Discov, 8, 2022
|
|
3RDH
 
 | | X-ray induced covalent inhibition of 14-3-3 | | 分子名称: | 14-3-3 protein zeta/delta, 4-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]benzoic acid, NICKEL (II) ION | | 著者 | Horton, J.R, Upadhyay, A.K, Fu, H, Cheng, X. | | 登録日 | 2011-04-01 | | 公開日 | 2011-09-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Discovery and structural characterization of a small molecule 14-3-3 protein-protein interaction inhibitor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4FLN
 
 | | Crystal structure of plant protease Deg2 | | 分子名称: | Protease Do-like 2, chloroplastic, Unknown peptide | | 著者 | Gong, W, Liu, L, Sun, R, Gao, F. | | 登録日 | 2012-06-15 | | 公開日 | 2012-09-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of Arabidopsis deg2 protein reveals an internal PDZ ligand locking the hexameric resting state.
J.Biol.Chem., 287, 2012
|
|
3SQJ
 
 | |
5ZZD
 
 | | Crystal structure of a protein from Aspergillus flavus | | 分子名称: | O-methyltransferase lepI, S-ADENOSYLMETHIONINE | | 著者 | Chang, Z.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2018-05-31 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of LepI, a multifunctional SAM-dependent enzyme which catalyzes pericyclic reactions in leporin biosynthesis.
Org.Biomol.Chem., 17, 2019
|
|
4FZW
 
 | | Crystal Structure of the PaaF-PaaG Hydratase-Isomerase Complex from E.coli | | 分子名称: | 1,2-epoxyphenylacetyl-CoA isomerase, 2,3-dehydroadipyl-CoA hydratase, GLYCEROL | | 著者 | Grishin, A.M, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | 登録日 | 2012-07-08 | | 公開日 | 2012-09-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Protein-Protein Interactions in the beta-Oxidation Part of the Phenylacetate
Utilization Pathway. Crystal Structure of the PaaF-PaaG Hydratase-Isomerase Complex
J.Biol.Chem., 287, 2012
|
|
6IEG
 
 | | Crystal structure of human MTR4 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Exosome RNA helicase MTR4, MAGNESIUM ION | | 著者 | Chen, J.Y, Yun, C.H. | | 登録日 | 2018-09-14 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.55 Å) | | 主引用文献 | NRDE2 negatively regulates exosome functions by inhibiting MTR4 recruitment and exosome interaction.
Genes Dev., 33, 2019
|
|
6M0J
 
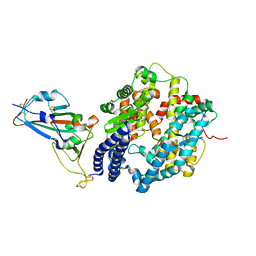 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain bound with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | 著者 | Wang, X, Lan, J, Ge, J, Yu, J, Shan, S. | | 登録日 | 2020-02-21 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor.
Nature, 581, 2020
|
|
7KK6
 
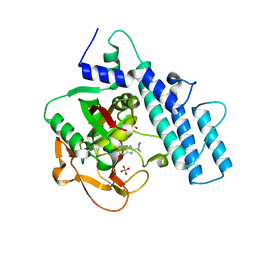 | |
7KKQ
 
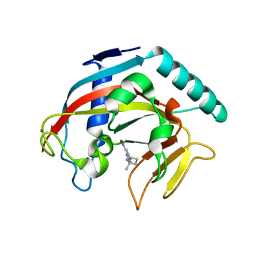 | |
7KK2
 
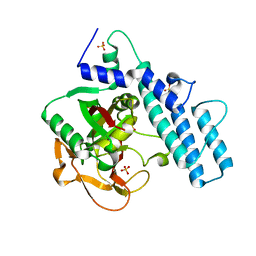 | |
5J8V
 
 | |
7KK5
 
 | |
7KKP
 
 | |
7KKO
 
 | |
7KKM
 
 | |
7BQY
 
 | | THE CRYSTAL STRUCTURE OF COVID-19 MAIN PROTEASE IN COMPLEX WITH AN INHIBITOR N3 at 1.7 angstrom | | 分子名称: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | 著者 | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | 登録日 | 2020-03-26 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
7KKN
 
 | | Structure of the catalytic domain of tankyrase 1 in complex with talazoparib | | 分子名称: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Poly [ADP-ribose] polymerase, ... | | 著者 | Gajiwala, K.S, Ryan, K. | | 登録日 | 2020-10-27 | | 公開日 | 2021-01-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KK4
 
 | |
